Henry Watkins
Building Models of Neurological Language
Jun 06, 2025Abstract:This report documents the development and evaluation of domain-specific language models for neurology. Initially focused on building a bespoke model, the project adapted to rapid advances in open-source and commercial medical LLMs, shifting toward leveraging retrieval-augmented generation (RAG) and representational models for secure, local deployment. Key contributions include the creation of neurology-specific datasets (case reports, QA sets, textbook-derived data), tools for multi-word expression extraction, and graph-based analyses of medical terminology. The project also produced scripts and Docker containers for local hosting. Performance metrics and graph community results are reported, with future possible work open for multimodal models using open-source architectures like phi-4.
Generalizable automated ischaemic stroke lesion segmentation with vision transformers
Feb 10, 2025
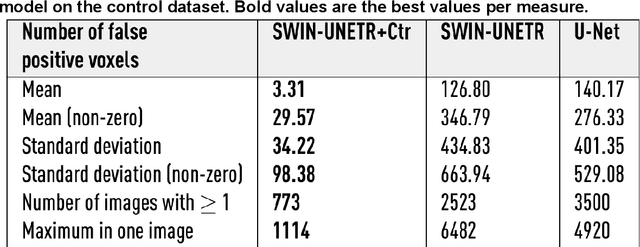
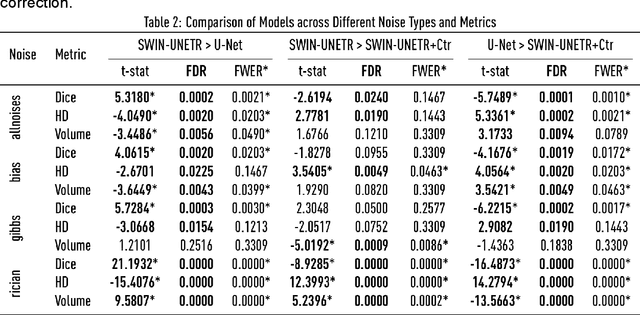
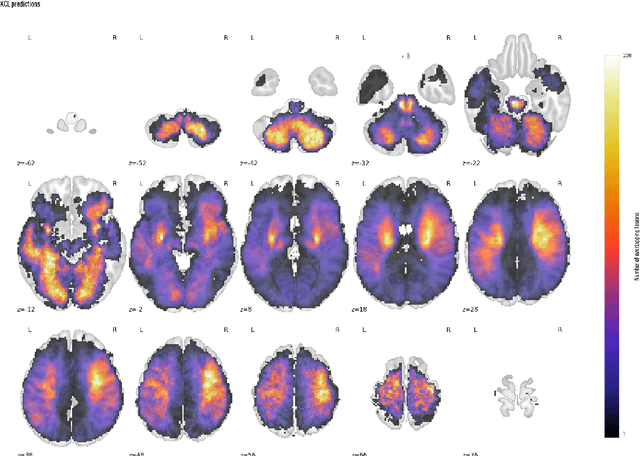
Abstract:Ischaemic stroke, a leading cause of death and disability, critically relies on neuroimaging for characterising the anatomical pattern of injury. Diffusion-weighted imaging (DWI) provides the highest expressivity in ischemic stroke but poses substantial challenges for automated lesion segmentation: susceptibility artefacts, morphological heterogeneity, age-related comorbidities, time-dependent signal dynamics, instrumental variability, and limited labelled data. Current U-Net-based models therefore underperform, a problem accentuated by inadequate evaluation metrics that focus on mean performance, neglecting anatomical, subpopulation, and acquisition-dependent variability. Here, we present a high-performance DWI lesion segmentation tool addressing these challenges through optimized vision transformer-based architectures, integration of 3563 annotated lesions from multi-site data, and algorithmic enhancements, achieving state-of-the-art results. We further propose a novel evaluative framework assessing model fidelity, equity (across demographics and lesion subtypes), anatomical precision, and robustness to instrumental variability, promoting clinical and research utility. This work advances stroke imaging by reconciling model expressivity with domain-specific challenges and redefining performance benchmarks to prioritize equity and generalizability, critical for personalized medicine and mechanistic research.
Compressed representation of brain genetic transcription
Oct 24, 2023Abstract:The architecture of the brain is too complex to be intuitively surveyable without the use of compressed representations that project its variation into a compact, navigable space. The task is especially challenging with high-dimensional data, such as gene expression, where the joint complexity of anatomical and transcriptional patterns demands maximum compression. Established practice is to use standard principal component analysis (PCA), whose computational felicity is offset by limited expressivity, especially at great compression ratios. Employing whole-brain, voxel-wise Allen Brain Atlas transcription data, here we systematically compare compressed representations based on the most widely supported linear and non-linear methods-PCA, kernel PCA, non-negative matrix factorization (NMF), t-stochastic neighbour embedding (t-SNE), uniform manifold approximation and projection (UMAP), and deep auto-encoding-quantifying reconstruction fidelity, anatomical coherence, and predictive utility with respect to signalling, microstructural, and metabolic targets. We show that deep auto-encoders yield superior representations across all metrics of performance and target domains, supporting their use as the reference standard for representing transcription patterns in the human brain.
An artificial intelligence natural language processing pipeline for information extraction in neuroradiology
Jul 21, 2021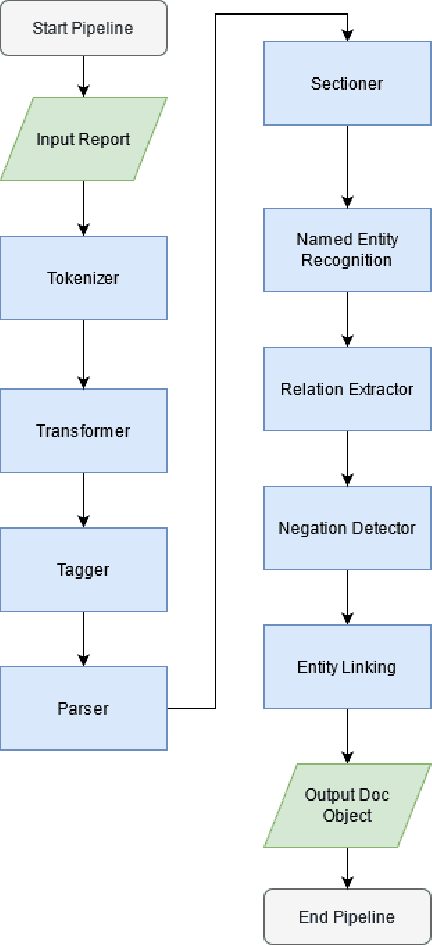
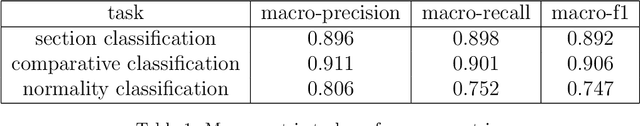


Abstract:The use of electronic health records in medical research is difficult because of the unstructured format. Extracting information within reports and summarising patient presentations in a way amenable to downstream analysis would be enormously beneficial for operational and clinical research. In this work we present a natural language processing pipeline for information extraction of radiological reports in neurology. Our pipeline uses a hybrid sequence of rule-based and artificial intelligence models to accurately extract and summarise neurological reports. We train and evaluate a custom language model on a corpus of 150000 radiological reports from National Hospital for Neurology and Neurosurgery, London MRI imaging. We also present results for standard NLP tasks on domain-specific neuroradiology datasets. We show our pipeline, called `neuroNLP', can reliably extract clinically relevant information from these reports, enabling downstream modelling of reports and associated imaging on a heretofore unprecedented scale.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge