Michel Thiebaut de Schotten
Conference Proceedings of the Inaugural Conference of the International Society for Tractography (IST 2025 Bordeaux)
Feb 12, 2026Abstract:This collection comprises the abstracts presented during poster, power pitch and oral sessions at the Inaugural Conference of the International Society for Tractography (IST Conference 2025), held in Bordeaux, France, from October 13-16, 2025. The conference was designed to foster meaningful exchange and collaboration between disparate fields. The overall focus was on advancing research, innovation, and community in the common fields of interest: neuroanatomy, tractography methods and scientific/clinical applications of tractography. The included abstracts cover the latest advancements in tractography, Diffusion MRI, and related fields including new work on; neurological and psychiatric disorders, deep brain stimulation targeting, and brain development. This landmark event brought together world-leading experts to discuss critical challenges and chart the future direction of the field.
Emerging-properties Mapping Using Spatial Embedding Statistics: EMUSES
Jun 20, 2024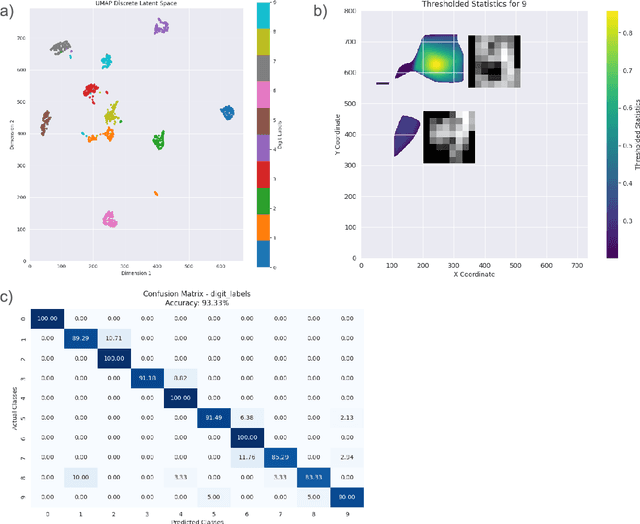
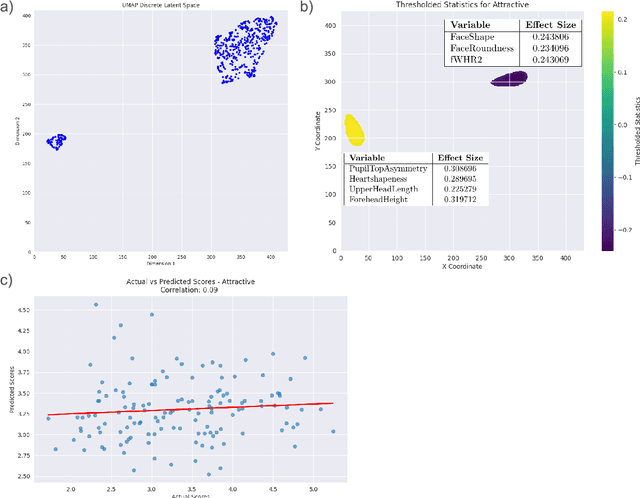
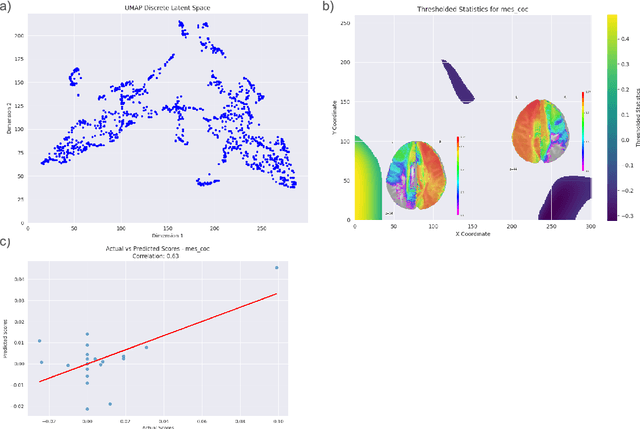
Abstract:Understanding complex phenomena often requires analyzing high-dimensional data to uncover emergent properties that arise from multifactorial interactions. Here, we present EMUSES (Emerging-properties Mapping Using Spatial Embedding Statistics), an innovative approach employing Uniform Manifold Approximation and Projection (UMAP) to create high-dimensional embeddings that reveal latent structures within data. EMUSES facilitates the exploration and prediction of emergent properties by statistically analyzing these latent spaces. Using three distinct datasets--a handwritten digits dataset from the National Institute of Standards and Technology (NIST, E. Alpaydin, 1998), the Chicago Face Database (Ma et al., 2015), and brain disconnection data post-stroke (Talozzi et al., 2023)--we demonstrate EMUSES' effectiveness in detecting and interpreting emergent properties. Our method not only predicts outcomes with high accuracy but also provides clear visualizations and statistical insights into the underlying interactions within the data. By bridging the gap between predictive accuracy and interpretability, EMUSES offers researchers a powerful tool to understand the multifactorial origins of complex phenomena.
Compressed representation of brain genetic transcription
Oct 24, 2023Abstract:The architecture of the brain is too complex to be intuitively surveyable without the use of compressed representations that project its variation into a compact, navigable space. The task is especially challenging with high-dimensional data, such as gene expression, where the joint complexity of anatomical and transcriptional patterns demands maximum compression. Established practice is to use standard principal component analysis (PCA), whose computational felicity is offset by limited expressivity, especially at great compression ratios. Employing whole-brain, voxel-wise Allen Brain Atlas transcription data, here we systematically compare compressed representations based on the most widely supported linear and non-linear methods-PCA, kernel PCA, non-negative matrix factorization (NMF), t-stochastic neighbour embedding (t-SNE), uniform manifold approximation and projection (UMAP), and deep auto-encoding-quantifying reconstruction fidelity, anatomical coherence, and predictive utility with respect to signalling, microstructural, and metabolic targets. We show that deep auto-encoders yield superior representations across all metrics of performance and target domains, supporting their use as the reference standard for representing transcription patterns in the human brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge