Danilo Bzdok
PARIETAL, NEUROSPIN
From Noise to Narrative: Tracing the Origins of Hallucinations in Transformers
Sep 08, 2025Abstract:As generative AI systems become competent and democratized in science, business, and government, deeper insight into their failure modes now poses an acute need. The occasional volatility in their behavior, such as the propensity of transformer models to hallucinate, impedes trust and adoption of emerging AI solutions in high-stakes areas. In the present work, we establish how and when hallucinations arise in pre-trained transformer models through concept representations captured by sparse autoencoders, under scenarios with experimentally controlled uncertainty in the input space. Our systematic experiments reveal that the number of semantic concepts used by the transformer model grows as the input information becomes increasingly unstructured. In the face of growing uncertainty in the input space, the transformer model becomes prone to activate coherent yet input-insensitive semantic features, leading to hallucinated output. At its extreme, for pure-noise inputs, we identify a wide variety of robustly triggered and meaningful concepts in the intermediate activations of pre-trained transformer models, whose functional integrity we confirm through targeted steering. We also show that hallucinations in the output of a transformer model can be reliably predicted from the concept patterns embedded in transformer layer activations. This collection of insights on transformer internal processing mechanics has immediate consequences for aligning AI models with human values, AI safety, opening the attack surface for potential adversarial attacks, and providing a basis for automatic quantification of a model's hallucination risk.
Prisma: An Open Source Toolkit for Mechanistic Interpretability in Vision and Video
Apr 28, 2025Abstract:Robust tooling and publicly available pre-trained models have helped drive recent advances in mechanistic interpretability for language models. However, similar progress in vision mechanistic interpretability has been hindered by the lack of accessible frameworks and pre-trained weights. We present Prisma (Access the codebase here: https://github.com/Prisma-Multimodal/ViT-Prisma), an open-source framework designed to accelerate vision mechanistic interpretability research, providing a unified toolkit for accessing 75+ vision and video transformers; support for sparse autoencoder (SAE), transcoder, and crosscoder training; a suite of 80+ pre-trained SAE weights; activation caching, circuit analysis tools, and visualization tools; and educational resources. Our analysis reveals surprising findings, including that effective vision SAEs can exhibit substantially lower sparsity patterns than language SAEs, and that in some instances, SAE reconstructions can decrease model loss. Prisma enables new research directions for understanding vision model internals while lowering barriers to entry in this emerging field.
Steering CLIP's vision transformer with sparse autoencoders
Apr 11, 2025



Abstract:While vision models are highly capable, their internal mechanisms remain poorly understood -- a challenge which sparse autoencoders (SAEs) have helped address in language, but which remains underexplored in vision. We address this gap by training SAEs on CLIP's vision transformer and uncover key differences between vision and language processing, including distinct sparsity patterns for SAEs trained across layers and token types. We then provide the first systematic analysis on the steerability of CLIP's vision transformer by introducing metrics to quantify how precisely SAE features can be steered to affect the model's output. We find that 10-15\% of neurons and features are steerable, with SAEs providing thousands more steerable features than the base model. Through targeted suppression of SAE features, we then demonstrate improved performance on three vision disentanglement tasks (CelebA, Waterbirds, and typographic attacks), finding optimal disentanglement in middle model layers, and achieving state-of-the-art performance on defense against typographic attacks.
ImageFlowNet: Forecasting Multiscale Trajectories of Disease Progression with Irregularly-Sampled Longitudinal Medical Images
Jun 20, 2024



Abstract:The forecasting of disease progression from images is a holy grail for clinical decision making. However, this task is complicated by the inherent high dimensionality, temporal sparsity and sampling irregularity in longitudinal image acquisitions. Existing methods often rely on extracting hand-crafted features and performing time-series analysis in this vector space, leading to a loss of rich spatial information within the images. To overcome these challenges, we introduce ImageFlowNet, a novel framework that learns latent-space flow fields that evolve multiscale representations in joint embedding spaces using neural ODEs and SDEs to model disease progression in the image domain. Notably, ImageFlowNet learns multiscale joint representation spaces by combining cohorts of patients together so that information can be transferred between the patient samples. The dynamics then provide plausible trajectories of progression, with the SDE providing alternative trajectories from the same starting point. We provide theoretical insights that support our formulation of ODEs, and motivate our regularizations involving high-level visual features, latent space organization, and trajectory smoothness. We then demonstrate ImageFlowNet's effectiveness through empirical evaluations on three longitudinal medical image datasets depicting progression in retinal geographic atrophy, multiple sclerosis, and glioblastoma.
Estimating Unknown Population Sizes Using the Hypergeometric Distribution
Feb 22, 2024



Abstract:The multivariate hypergeometric distribution describes sampling without replacement from a discrete population of elements divided into multiple categories. Addressing a gap in the literature, we tackle the challenge of estimating discrete distributions when both the total population size and the sizes of its constituent categories are unknown. Here, we propose a novel solution using the hypergeometric likelihood to solve this estimation challenge, even in the presence of severe under-sampling. We develop our approach to account for a data generating process where the ground-truth is a mixture of distributions conditional on a continuous latent variable, such as with collaborative filtering, using the variational autoencoder framework. Empirical data simulation demonstrates that our method outperforms other likelihood functions used to model count data, both in terms of accuracy of population size estimate and in its ability to learn an informative latent space. We demonstrate our method's versatility through applications in NLP, by inferring and estimating the complexity of latent vocabularies in text excerpts, and in biology, by accurately recovering the true number of gene transcripts from sparse single-cell genomics data.
The Uncanny Valley: A Comprehensive Analysis of Diffusion Models
Feb 20, 2024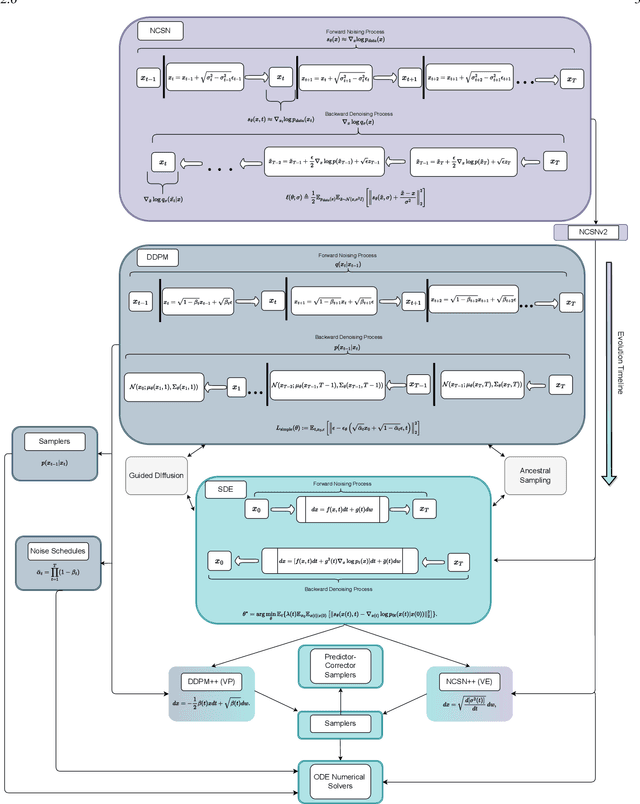
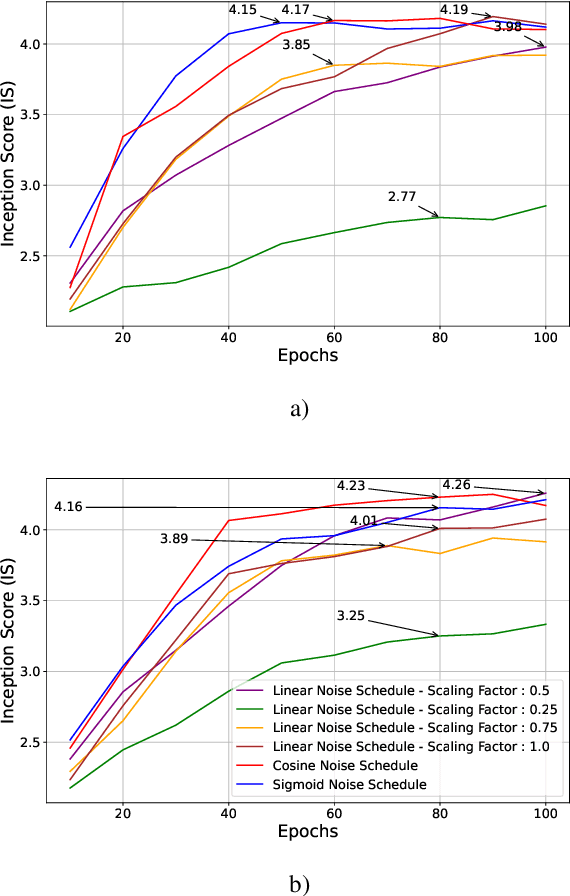
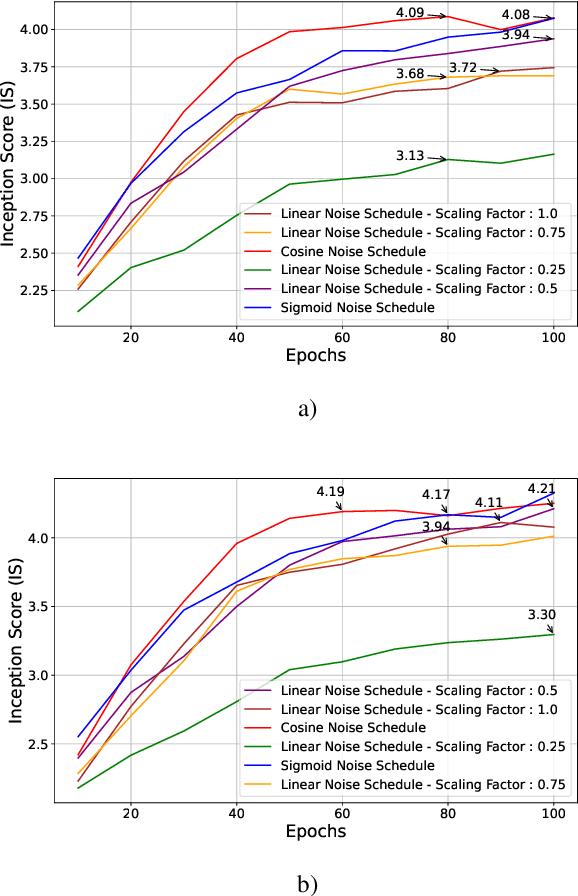
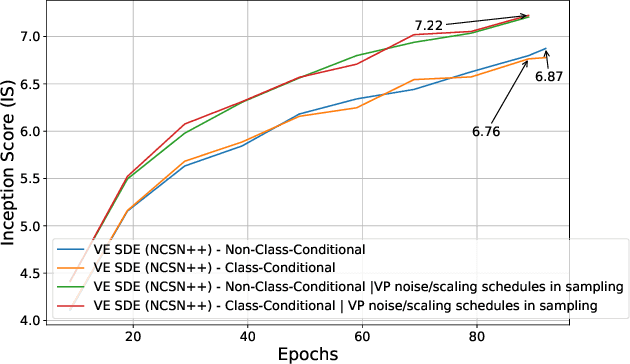
Abstract:Through Diffusion Models (DMs), we have made significant advances in generating high-quality images. Our exploration of these models delves deeply into their core operational principles by systematically investigating key aspects across various DM architectures: i) noise schedules, ii) samplers, and iii) guidance. Our comprehensive examination of these models sheds light on their hidden fundamental mechanisms, revealing the concealed foundational elements that are essential for their effectiveness. Our analyses emphasize the hidden key factors that determine model performance, offering insights that contribute to the advancement of DMs. Past findings show that the configuration of noise schedules, samplers, and guidance is vital to the quality of generated images; however, models reach a stable level of quality across different configurations at a remarkably similar point, revealing that the decisive factors for optimal performance predominantly reside in the diffusion process dynamics and the structural design of the model's network, rather than the specifics of configuration details. Our comparative analysis reveals that Denoising Diffusion Probabilistic Model (DDPM)-based diffusion dynamics consistently outperform the Noise Conditioned Score Network (NCSN)-based ones, not only when evaluated in their original forms but also when continuous through Stochastic Differential Equation (SDE)-based implementations.
Representational Ethical Model Calibration
Jul 25, 2022



Abstract:Equity is widely held to be fundamental to the ethics of healthcare. In the context of clinical decision-making, it rests on the comparative fidelity of the intelligence -- evidence-based or intuitive -- guiding the management of each individual patient. Though brought to recent attention by the individuating power of contemporary machine learning, such epistemic equity arises in the context of any decision guidance, whether traditional or innovative. Yet no general framework for its quantification, let alone assurance, currently exists. Here we formulate epistemic equity in terms of model fidelity evaluated over learnt multi-dimensional representations of identity crafted to maximise the captured diversity of the population, introducing a comprehensive framework for Representational Ethical Model Calibration. We demonstrate use of the framework on large-scale multimodal data from UK Biobank to derive diverse representations of the population, quantify model performance, and institute responsive remediation. We offer our approach as a principled solution to quantifying and assuring epistemic equity in healthcare, with applications across the research, clinical, and regulatory domains.
Orientation and Context Entangled Network for Retinal Vessel Segmentation
Jul 23, 2022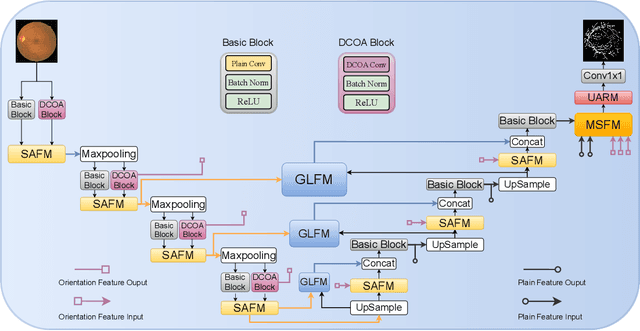
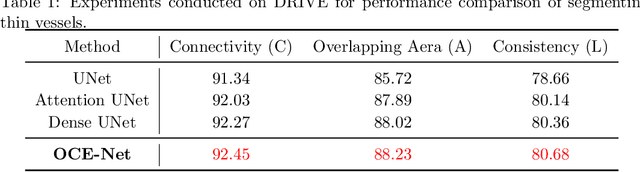
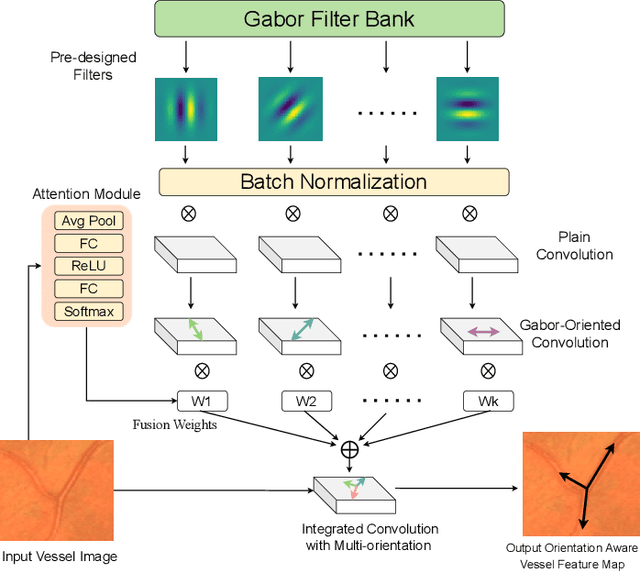
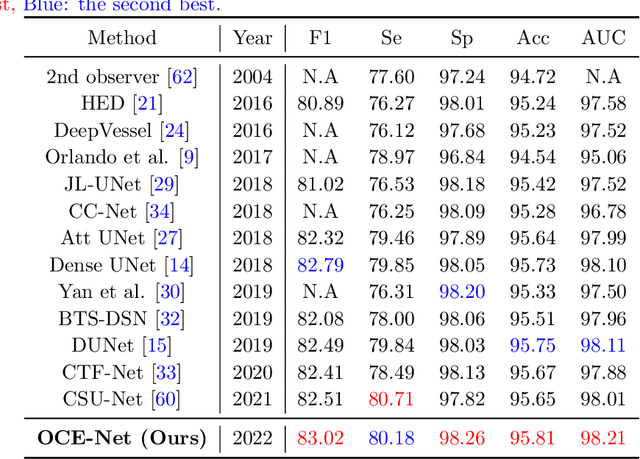
Abstract:Most of the existing deep learning based methods for vessel segmentation neglect two important aspects of retinal vessels, one is the orientation information of vessels, and the other is the contextual information of the whole fundus region. In this paper, we propose a robust Orientation and Context Entangled Network (denoted as OCE-Net), which has the capability of extracting complex orientation and context information of the blood vessels. To achieve complex orientation aware, a Dynamic Complex Orientation Aware Convolution (DCOA Conv) is proposed to extract complex vessels with multiple orientations for improving the vessel continuity. To simultaneously capture the global context information and emphasize the important local information, a Global and Local Fusion Module (GLFM) is developed to simultaneously model the long-range dependency of vessels and focus sufficient attention on local thin vessels. A novel Orientation and Context Entangled Non-local (OCE-NL) module is proposed to entangle the orientation and context information together. In addition, an Unbalanced Attention Refining Module (UARM) is proposed to deal with the unbalanced pixel numbers of background, thick and thin vessels. Extensive experiments were performed on several commonly used datasets (DRIVE, STARE and CHASEDB1) and some more challenging datasets (AV-WIDE, UoA-DR, RFMiD and UK Biobank). The ablation study shows that the proposed method achieves promising performance on maintaining the continuity of thin vessels and the comparative experiments demonstrate that our OCE-Net can achieve state-of-the-art performance on retinal vessel segmentation.
Label scarcity in biomedicine: Data-rich latent factor discovery enhances phenotype prediction
Oct 12, 2021


Abstract:High-quality data accumulation is now becoming ubiquitous in the health domain. There is increasing opportunity to exploit rich data from normal subjects to improve supervised estimators in specific diseases with notorious data scarcity. We demonstrate that low-dimensional embedding spaces can be derived from the UK Biobank population dataset and used to enhance data-scarce prediction of health indicators, lifestyle and demographic characteristics. Phenotype predictions facilitated by Variational Autoencoder manifolds typically scaled better with increasing unlabeled data than dimensionality reduction by PCA or Isomap. Performances gains from semisupervison approaches will probably become an important ingredient for various medical data science applications.
Clusters in Explanation Space: Inferring disease subtypes from model explanations
Dec 18, 2019

Abstract:Identification of disease subtypes and corresponding biomarkers can substantially improve clinical diagnosis and treatment selection. Discovering these subtypes in noisy, high dimensional biomedical data is often impossible for humans and challenging for machines. We introduce a new approach to facilitate the discovery of disease subtypes: Instead of analyzing the original data, we train a diagnostic classifier (healthy vs. diseased) and extract instance-wise explanations for the classifier's decisions. The distribution of instances in the explanation space of our diagnostic classifier amplifies the different reasons for belonging to the same class - resulting in a representation that is uniquely useful for discovering latent subtypes. We compare our ability to recover subtypes via cluster analysis on model explanations to classical cluster analysis on the original data. In multiple datasets with known ground-truth subclasses, most compellingly on UK Biobank brain imaging data and transcriptome data from the Cancer Genome Atlas, we show that cluster analysis on model explanations substantially outperforms the classical approach. While we believe clustering in explanation space to be particularly valuable for inferring disease subtypes, the method is more general and applicable to any kind of sub-type identification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge