Janaina Mourao-Miranda
Interpretable Generative and Discriminative Learning for Multimodal and Incomplete Clinical Data
Oct 10, 2025Abstract:Real-world clinical problems are often characterized by multimodal data, usually associated with incomplete views and limited sample sizes in their cohorts, posing significant limitations for machine learning algorithms. In this work, we propose a Bayesian approach designed to efficiently handle these challenges while providing interpretable solutions. Our approach integrates (1) a generative formulation to capture cross-view relationships with a semi-supervised strategy, and (2) a discriminative task-oriented formulation to identify relevant information for specific downstream objectives. This dual generative-discriminative formulation offers both general understanding and task-specific insights; thus, it provides an automatic imputation of the missing views while enabling robust inference across different data sources. The potential of this approach becomes evident when applied to the multimodal clinical data, where our algorithm is able to capture and disentangle the complex interactions among biological, psychological, and sociodemographic modalities.
Identifying latent disease factors differently expressed in patient subgroups using group factor analysis
Oct 10, 2024



Abstract:In this study, we propose a novel approach to uncover subgroup-specific and subgroup-common latent factors addressing the challenges posed by the heterogeneity of neurological and mental disorders, which hinder disease understanding, treatment development, and outcome prediction. The proposed approach, sparse Group Factor Analysis (GFA) with regularised horseshoe priors, was implemented with probabilistic programming and can uncover associations (or latent factors) among multiple data modalities differentially expressed in sample subgroups. Synthetic data experiments showed the robustness of our sparse GFA by correctly inferring latent factors and model parameters. When applied to the Genetic Frontotemporal Dementia Initiative (GENFI) dataset, which comprises patients with frontotemporal dementia (FTD) with genetically defined subgroups, the sparse GFA identified latent disease factors differentially expressed across the subgroups, distinguishing between "subgroup-specific" latent factors within homogeneous groups and "subgroup common" latent factors shared across subgroups. The latent disease factors captured associations between brain structure and non-imaging variables (i.e., questionnaires assessing behaviour and disease severity) across the different genetic subgroups, offering insights into disease profiles. Importantly, two latent factors were more pronounced in the two more homogeneous FTD patient subgroups (progranulin (GRN) and microtubule-associated protein tau (MAPT) mutation), showcasing the method's ability to reveal subgroup-specific characteristics. These findings underscore the potential of sparse GFA for integrating multiple data modalities and identifying interpretable latent disease factors that can improve the characterization and stratification of patients with neurological and mental health disorders.
TransductGAN: a Transductive Adversarial Model for Novelty Detection
Mar 30, 2022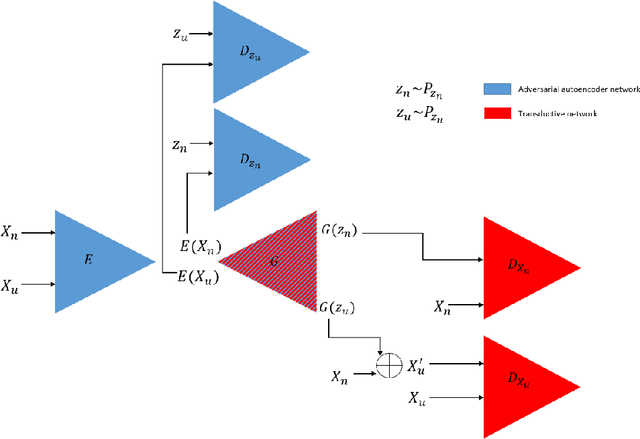

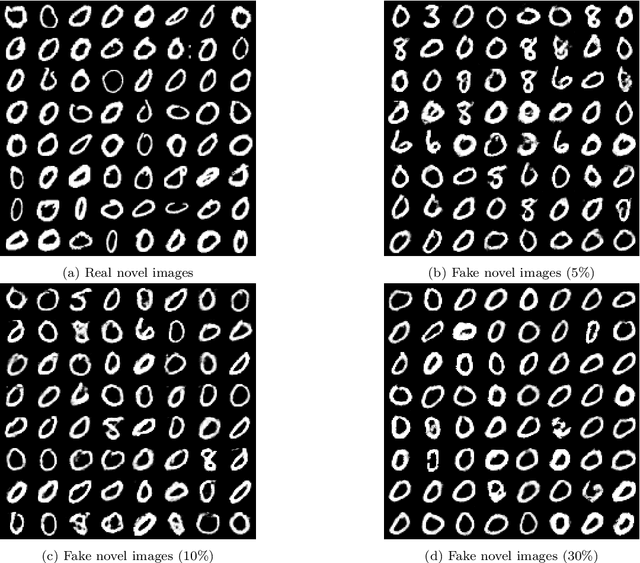
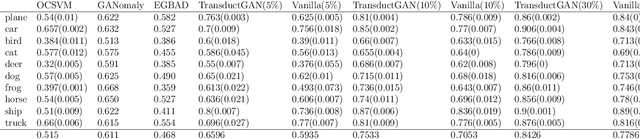
Abstract:Novelty detection, a widely studied problem in machine learning, is the problem of detecting a novel class of data that has not been previously observed. A common setting for novelty detection is inductive whereby only examples of the negative class are available during training time. Transductive novelty detection on the other hand has only witnessed a recent surge in interest, it not only makes use of the negative class during training but also incorporates the (unlabeled) test set to detect novel examples. Several studies have emerged under the transductive setting umbrella that have demonstrated its advantage over its inductive counterpart. Depending on the assumptions about the data, these methods go by different names (e.g. transductive novelty detection, semi-supervised novelty detection, positive-unlabeled learning, out-of-distribution detection). With the use of generative adversarial networks (GAN), a segment of those studies have adopted a transductive setup in order to learn how to generate examples of the novel class. In this study, we propose TransductGAN, a transductive generative adversarial network that attempts to learn how to generate image examples from both the novel and negative classes by using a mixture of two Gaussians in the latent space. It achieves that by incorporating an adversarial autoencoder with a GAN network, the ability to generate examples of novel data points offers not only a visual representation of novelties, but also overcomes the hurdle faced by many inductive methods of how to tune the model hyperparameters at the decision rule level. Our model has shown superior performance over state-of-the-art inductive and transductive methods. Our study is fully reproducible with the code available publicly.
A hierarchical Bayesian model to find brain-behaviour associations in incomplete data sets
Mar 11, 2021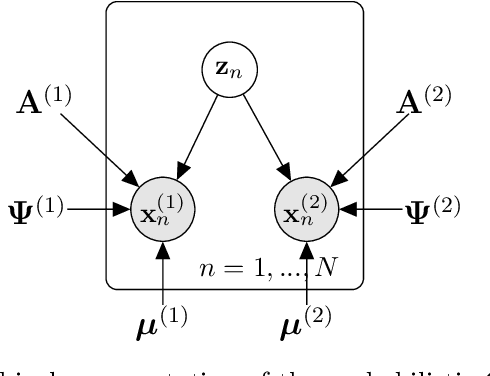

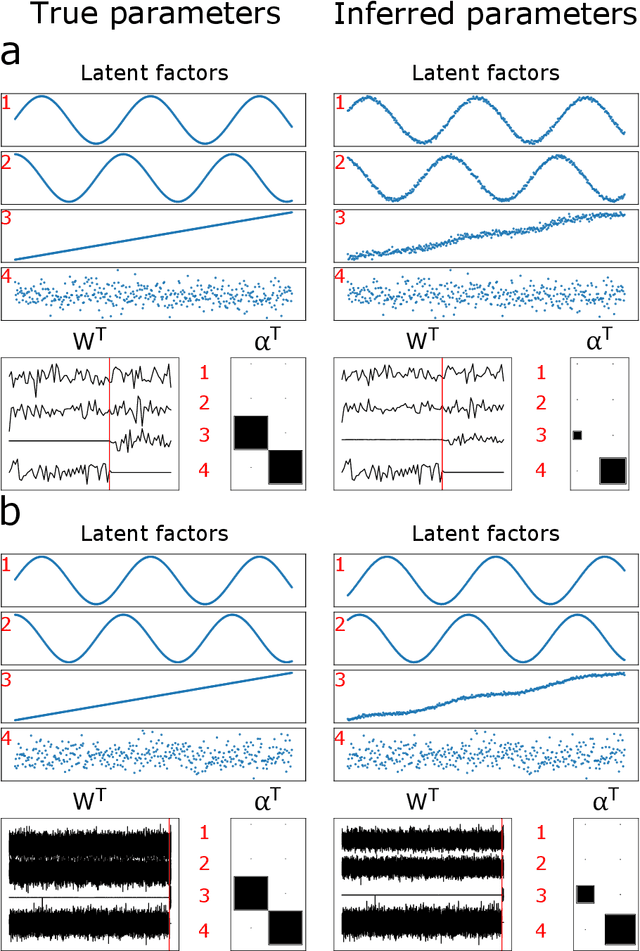

Abstract:Canonical Correlation Analysis (CCA) and its regularised versions have been widely used in the neuroimaging community to uncover multivariate associations between two data modalities (e.g., brain imaging and behaviour). However, these methods have inherent limitations: (1) statistical inferences about the associations are often not robust; (2) the associations within each data modality are not modelled; (3) missing values need to be imputed or removed. Group Factor Analysis (GFA) is a hierarchical model that addresses the first two limitations by providing Bayesian inference and modelling modality-specific associations. Here, we propose an extension of GFA that handles missing data, and highlight that GFA can be used as a predictive model. We applied GFA to synthetic and real data consisting of brain connectivity and non-imaging measures from the Human Connectome Project (HCP). In synthetic data, GFA uncovered the underlying shared and specific factors and predicted correctly the non-observed data modalities in complete and incomplete data sets. In the HCP data, we identified four relevant shared factors, capturing associations between mood, alcohol and drug use, cognition, demographics and psychopathological measures and the default mode, frontoparietal control, dorsal and ventral networks and insula, as well as two factors describing associations within brain connectivity. In addition, GFA predicted a set of non-imaging measures from brain connectivity. These findings were consistent in complete and incomplete data sets, and replicated previous findings in the literature. GFA is a promising tool that can be used to uncover associations between and within multiple data modalities in benchmark datasets (such as, HCP), and easily extended to more complex models to solve more challenging tasks.
Finding the needle in high-dimensional haystack: A tutorial on canonical correlation analysis
Dec 06, 2018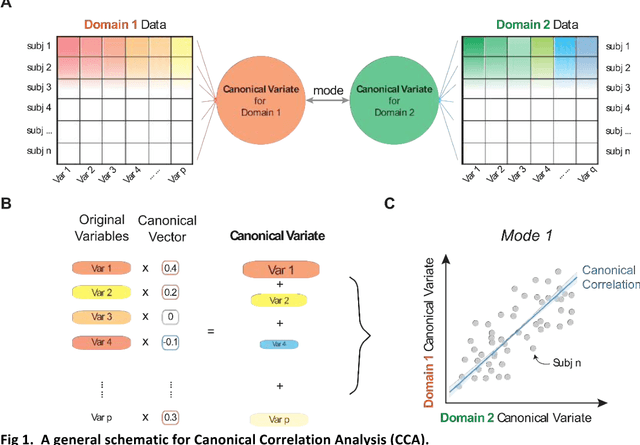
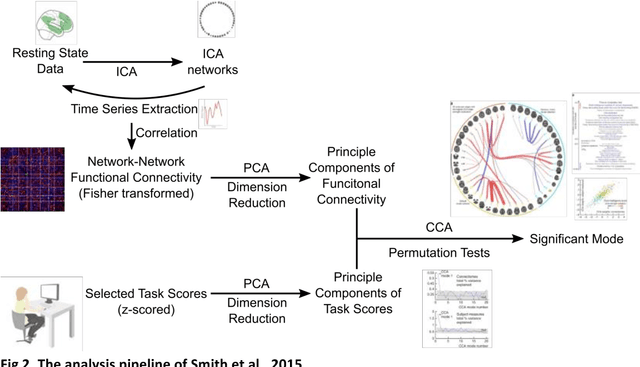
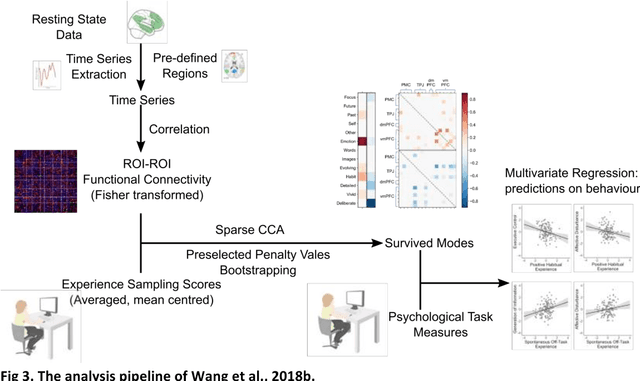
Abstract:Since the beginning of the 21st century, the size, breadth, and granularity of data in biology and medicine has grown rapidly. In the example of neuroscience, studies with thousands of subjects are becoming more common, which provide extensive phenotyping on the behavioral, neural, and genomic level with hundreds of variables. The complexity of such big data repositories offer new opportunities and pose new challenges to investigate brain, cognition, and disease. Canonical correlation analysis (CCA) is a prototypical family of methods for wrestling with and harvesting insight from such rich datasets. This doubly-multivariate tool can simultaneously consider two variable sets from different modalities to uncover essential hidden associations. Our primer discusses the rationale, promises, and pitfalls of CCA in biomedicine.
Interpreting weight maps in terms of cognitive or clinical neuroscience: nonsense?
Apr 30, 2018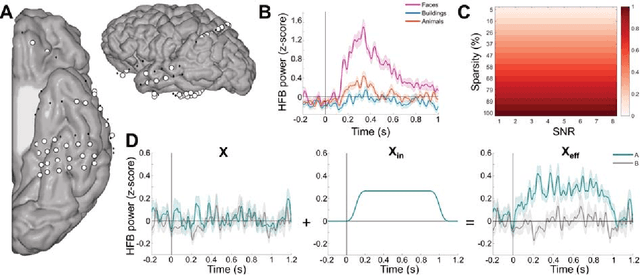
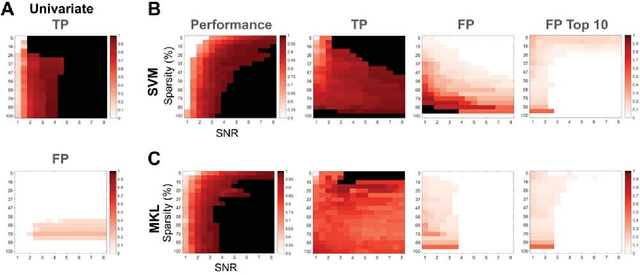
Abstract:Since machine learning models have been applied to neuroimaging data, researchers have drawn conclusions from the derived weight maps. In particular, weight maps of classifiers between two conditions are often described as a proxy for the underlying signal differences between the conditions. Recent studies have however suggested that such weight maps could not reliably recover the source of the neural signals and even led to false positives (FP). In this work, we used semi-simulated data from ElectroCorticoGraphy (ECoG) to investigate how the signal-to-noise ratio and sparsity of the neural signal affect the similarity between signal and weights. We show that not all cases produce FP and that it is unlikely for FP features to have a high weight in most cases.
* conference article
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge