John C. van Swieten
for the Alzheimer's Disease Neuroimaging Initiative, on behalf of the Parelsnoer Neurodegenerative Diseases study group
Identifying latent disease factors differently expressed in patient subgroups using group factor analysis
Oct 10, 2024



Abstract:In this study, we propose a novel approach to uncover subgroup-specific and subgroup-common latent factors addressing the challenges posed by the heterogeneity of neurological and mental disorders, which hinder disease understanding, treatment development, and outcome prediction. The proposed approach, sparse Group Factor Analysis (GFA) with regularised horseshoe priors, was implemented with probabilistic programming and can uncover associations (or latent factors) among multiple data modalities differentially expressed in sample subgroups. Synthetic data experiments showed the robustness of our sparse GFA by correctly inferring latent factors and model parameters. When applied to the Genetic Frontotemporal Dementia Initiative (GENFI) dataset, which comprises patients with frontotemporal dementia (FTD) with genetically defined subgroups, the sparse GFA identified latent disease factors differentially expressed across the subgroups, distinguishing between "subgroup-specific" latent factors within homogeneous groups and "subgroup common" latent factors shared across subgroups. The latent disease factors captured associations between brain structure and non-imaging variables (i.e., questionnaires assessing behaviour and disease severity) across the different genetic subgroups, offering insights into disease profiles. Importantly, two latent factors were more pronounced in the two more homogeneous FTD patient subgroups (progranulin (GRN) and microtubule-associated protein tau (MAPT) mutation), showcasing the method's ability to reveal subgroup-specific characteristics. These findings underscore the potential of sparse GFA for integrating multiple data modalities and identifying interpretable latent disease factors that can improve the characterization and stratification of patients with neurological and mental health disorders.
Cross-Cohort Generalizability of Deep and Conventional Machine Learning for MRI-based Diagnosis and Prediction of Alzheimer's Disease
Dec 16, 2020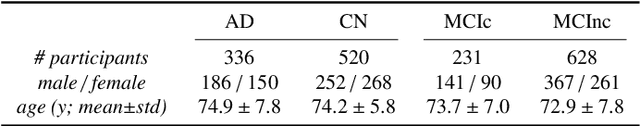
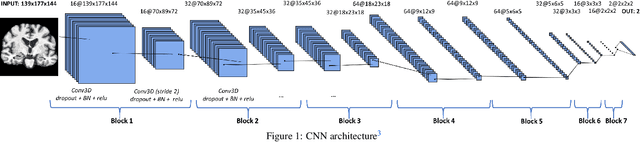
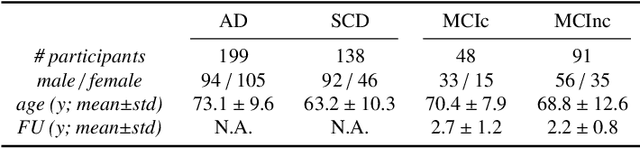
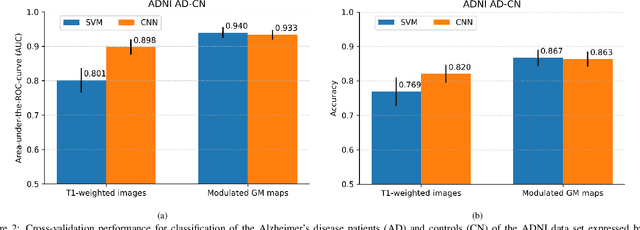
Abstract:This work validates the generalizability of MRI-based classification of Alzheimer's disease (AD) patients and controls (CN) to an external data set and to the task of prediction of conversion to AD in individuals with mild cognitive impairment (MCI). We used a conventional support vector machine (SVM) and a deep convolutional neural network (CNN) approach based on structural MRI scans that underwent either minimal pre-processing or more extensive pre-processing into modulated gray matter (GM) maps. Classifiers were optimized and evaluated using cross-validation in the ADNI (334 AD, 520 CN). Trained classifiers were subsequently applied to predict conversion to AD in ADNI MCI patients (231 converters, 628 non-converters) and in the independent Health-RI Parelsnoer data set. From this multi-center study representing a tertiary memory clinic population, we included 199 AD patients, 139 participants with subjective cognitive decline, 48 MCI patients converting to dementia, and 91 MCI patients who did not convert to dementia. AD-CN classification based on modulated GM maps resulted in a similar AUC for SVM (0.940) and CNN (0.933). Application to conversion prediction in MCI yielded significantly higher performance for SVM (0.756) than for CNN (0.742). In external validation, performance was slightly decreased. For AD-CN, it again gave similar AUCs for SVM (0.896) and CNN (0.876). For prediction in MCI, performances decreased for both SVM (0.665) and CNN (0.702). Both with SVM and CNN, classification based on modulated GM maps significantly outperformed classification based on minimally processed images. Deep and conventional classifiers performed equally well for AD classification and their performance decreased only slightly when applied to the external cohort. We expect that this work on external validation contributes towards translation of machine learning to clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge