Alexander Tong
FALCON: Few-step Accurate Likelihoods for Continuous Flows
Dec 10, 2025

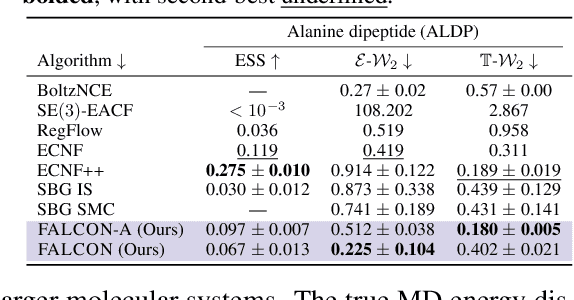

Abstract:Scalable sampling of molecular states in thermodynamic equilibrium is a long-standing challenge in statistical physics. Boltzmann Generators tackle this problem by pairing a generative model, capable of exact likelihood computation, with importance sampling to obtain consistent samples under the target distribution. Current Boltzmann Generators primarily use continuous normalizing flows (CNFs) trained with flow matching for efficient training of powerful models. However, likelihood calculation for these models is extremely costly, requiring thousands of function evaluations per sample, severely limiting their adoption. In this work, we propose Few-step Accurate Likelihoods for Continuous Flows (FALCON), a method which allows for few-step sampling with a likelihood accurate enough for importance sampling applications by introducing a hybrid training objective that encourages invertibility. We show FALCON outperforms state-of-the-art normalizing flow models for molecular Boltzmann sampling and is two orders of magnitude faster than the equivalently performing CNF model.
Curly Flow Matching for Learning Non-gradient Field Dynamics
Oct 30, 2025Abstract:Modeling the transport dynamics of natural processes from population-level observations is a ubiquitous problem in the natural sciences. Such models rely on key assumptions about the underlying process in order to enable faithful learning of governing dynamics that mimic the actual system behavior. The de facto assumption in current approaches relies on the principle of least action that results in gradient field dynamics and leads to trajectories minimizing an energy functional between two probability measures. However, many real-world systems, such as cell cycles in single-cell RNA, are known to exhibit non-gradient, periodic behavior, which fundamentally cannot be captured by current state-of-the-art methods such as flow and bridge matching. In this paper, we introduce Curly Flow Matching (Curly-FM), a novel approach that is capable of learning non-gradient field dynamics by designing and solving a Schr\"odinger bridge problem with a non-zero drift reference process -- in stark contrast to typical zero-drift reference processes -- which is constructed using inferred velocities in addition to population snapshot data. We showcase Curly-FM by solving the trajectory inference problems for single cells, computational fluid dynamics, and ocean currents with approximate velocities. We demonstrate that Curly-FM can learn trajectories that better match both the reference process and population marginals. Curly-FM expands flow matching models beyond the modeling of populations and towards the modeling of known periodic behavior in physical systems. Our code repository is accessible at: https://github.com/kpetrovicc/curly-flow-matching.git
Amortized Sampling with Transferable Normalizing Flows
Aug 25, 2025Abstract:Efficient equilibrium sampling of molecular conformations remains a core challenge in computational chemistry and statistical inference. Classical approaches such as molecular dynamics or Markov chain Monte Carlo inherently lack amortization; the computational cost of sampling must be paid in-full for each system of interest. The widespread success of generative models has inspired interest into overcoming this limitation through learning sampling algorithms. Despite performing on par with conventional methods when trained on a single system, learned samplers have so far demonstrated limited ability to transfer across systems. We prove that deep learning enables the design of scalable and transferable samplers by introducing Prose, a 280 million parameter all-atom transferable normalizing flow trained on a corpus of peptide molecular dynamics trajectories up to 8 residues in length. Prose draws zero-shot uncorrelated proposal samples for arbitrary peptide systems, achieving the previously intractable transferability across sequence length, whilst retaining the efficient likelihood evaluation of normalizing flows. Through extensive empirical evaluation we demonstrate the efficacy of Prose as a proposal for a variety of sampling algorithms, finding a simple importance sampling-based finetuning procedure to achieve superior performance to established methods such as sequential Monte Carlo on unseen tetrapeptides. We open-source the Prose codebase, model weights, and training dataset, to further stimulate research into amortized sampling methods and finetuning objectives.
Branched Schrödinger Bridge Matching
Jun 10, 2025Abstract:Predicting the intermediate trajectories between an initial and target distribution is a central problem in generative modeling. Existing approaches, such as flow matching and Schr\"odinger Bridge Matching, effectively learn mappings between two distributions by modeling a single stochastic path. However, these methods are inherently limited to unimodal transitions and cannot capture branched or divergent evolution from a common origin to multiple distinct outcomes. To address this, we introduce Branched Schr\"odinger Bridge Matching (BranchSBM), a novel framework that learns branched Schr\"odinger bridges. BranchSBM parameterizes multiple time-dependent velocity fields and growth processes, enabling the representation of population-level divergence into multiple terminal distributions. We show that BranchSBM is not only more expressive but also essential for tasks involving multi-path surface navigation, modeling cell fate bifurcations from homogeneous progenitor states, and simulating diverging cellular responses to perturbations.
Gumbel-Softmax Flow Matching with Straight-Through Guidance for Controllable Biological Sequence Generation
Mar 21, 2025Abstract:Flow matching in the continuous simplex has emerged as a promising strategy for DNA sequence design, but struggles to scale to higher simplex dimensions required for peptide and protein generation. We introduce Gumbel-Softmax Flow and Score Matching, a generative framework on the simplex based on a novel Gumbel-Softmax interpolant with a time-dependent temperature. Using this interpolant, we introduce Gumbel-Softmax Flow Matching by deriving a parameterized velocity field that transports from smooth categorical distributions to distributions concentrated at a single vertex of the simplex. We alternatively present Gumbel-Softmax Score Matching which learns to regress the gradient of the probability density. Our framework enables high-quality, diverse generation and scales efficiently to higher-dimensional simplices. To enable training-free guidance, we propose Straight-Through Guided Flows (STGFlow), a classifier-based guidance method that leverages straight-through estimators to steer the unconditional velocity field toward optimal vertices of the simplex. STGFlow enables efficient inference-time guidance using classifiers pre-trained on clean sequences, and can be used with any discrete flow method. Together, these components form a robust framework for controllable de novo sequence generation. We demonstrate state-of-the-art performance in conditional DNA promoter design, sequence-only protein generation, and target-binding peptide design for rare disease treatment.
Feynman-Kac Correctors in Diffusion: Annealing, Guidance, and Product of Experts
Mar 04, 2025Abstract:While score-based generative models are the model of choice across diverse domains, there are limited tools available for controlling inference-time behavior in a principled manner, e.g. for composing multiple pretrained models. Existing classifier-free guidance methods use a simple heuristic to mix conditional and unconditional scores to approximately sample from conditional distributions. However, such methods do not approximate the intermediate distributions, necessitating additional 'corrector' steps. In this work, we provide an efficient and principled method for sampling from a sequence of annealed, geometric-averaged, or product distributions derived from pretrained score-based models. We derive a weighted simulation scheme which we call Feynman-Kac Correctors (FKCs) based on the celebrated Feynman-Kac formula by carefully accounting for terms in the appropriate partial differential equations (PDEs). To simulate these PDEs, we propose Sequential Monte Carlo (SMC) resampling algorithms that leverage inference-time scaling to improve sampling quality. We empirically demonstrate the utility of our methods by proposing amortized sampling via inference-time temperature annealing, improving multi-objective molecule generation using pretrained models, and improving classifier-free guidance for text-to-image generation. Our code is available at https://github.com/martaskrt/fkc-diffusion.
Scalable Equilibrium Sampling with Sequential Boltzmann Generators
Feb 25, 2025

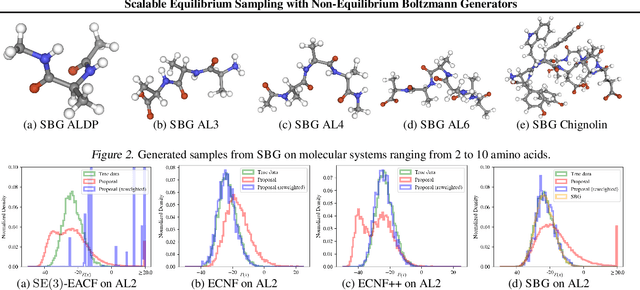

Abstract:Scalable sampling of molecular states in thermodynamic equilibrium is a long-standing challenge in statistical physics. Boltzmann generators tackle this problem by pairing powerful normalizing flows with importance sampling to obtain statistically independent samples under the target distribution. In this paper, we extend the Boltzmann generator framework and introduce Sequential Boltzmann generators (SBG) with two key improvements. The first is a highly efficient non-equivariant Transformer-based normalizing flow operating directly on all-atom Cartesian coordinates. In contrast to equivariant continuous flows of prior methods, we leverage exactly invertible non-equivariant architectures which are highly efficient both during sample generation and likelihood computation. As a result, this unlocks more sophisticated inference strategies beyond standard importance sampling. More precisely, as a second key improvement we perform inference-time scaling of flow samples using annealed Langevin dynamics which transports samples toward the target distribution leading to lower variance (annealed) importance weights which enable higher fidelity resampling with sequential Monte Carlo. SBG achieves state-of-the-art performance w.r.t. all metrics on molecular systems, demonstrating the first equilibrium sampling in Cartesian coordinates of tri, tetra, and hexapeptides that were so far intractable for prior Boltzmann generators.
Path Planning for Masked Diffusion Model Sampling
Feb 05, 2025Abstract:In this paper, we investigate how the order in which tokens are unmasked during masked diffusion models (MDMs) inference affects generative quality. We derive an expanded evidence lower bound (ELBO) that introduces a planner, responsible for selecting which tokens to unmask at each step. Our analysis suggests that alternative unmasking strategies can improve generative performance. Based on these insights, we propose Path Planning (P2), a sampling framework that leverages pre-trained BERT or the denoiser itself to guide unmasking decisions. P2 generalizes all known MDM sampling strategies and enables significant improvements across diverse domains including language generation (in-context learning, code generation, story infilling, mathematical reasoning, reverse curse correction) and biological sequence generation (protein and RNA sequences).
The Superposition of Diffusion Models Using the Itô Density Estimator
Dec 23, 2024Abstract:The Cambrian explosion of easily accessible pre-trained diffusion models suggests a demand for methods that combine multiple different pre-trained diffusion models without incurring the significant computational burden of re-training a larger combined model. In this paper, we cast the problem of combining multiple pre-trained diffusion models at the generation stage under a novel proposed framework termed superposition. Theoretically, we derive superposition from rigorous first principles stemming from the celebrated continuity equation and design two novel algorithms tailor-made for combining diffusion models in SuperDiff. SuperDiff leverages a new scalable It\^o density estimator for the log likelihood of the diffusion SDE which incurs no additional overhead compared to the well-known Hutchinson's estimator needed for divergence calculations. We demonstrate that SuperDiff is scalable to large pre-trained diffusion models as superposition is performed solely through composition during inference, and also enjoys painless implementation as it combines different pre-trained vector fields through an automated re-weighting scheme. Notably, we show that SuperDiff is efficient during inference time, and mimics traditional composition operators such as the logical OR and the logical AND. We empirically demonstrate the utility of using SuperDiff for generating more diverse images on CIFAR-10, more faithful prompt conditioned image editing using Stable Diffusion, and improved unconditional de novo structure design of proteins. https://github.com/necludov/super-diffusion
Trajectory Flow Matching with Applications to Clinical Time Series Modeling
Oct 28, 2024Abstract:Modeling stochastic and irregularly sampled time series is a challenging problem found in a wide range of applications, especially in medicine. Neural stochastic differential equations (Neural SDEs) are an attractive modeling technique for this problem, which parameterize the drift and diffusion terms of an SDE with neural networks. However, current algorithms for training Neural SDEs require backpropagation through the SDE dynamics, greatly limiting their scalability and stability. To address this, we propose Trajectory Flow Matching (TFM), which trains a Neural SDE in a simulation-free manner, bypassing backpropagation through the dynamics. TFM leverages the flow matching technique from generative modeling to model time series. In this work we first establish necessary conditions for TFM to learn time series data. Next, we present a reparameterization trick which improves training stability. Finally, we adapt TFM to the clinical time series setting, demonstrating improved performance on three clinical time series datasets both in terms of absolute performance and uncertainty prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge