Zhangzhi Peng
Steering Masked Discrete Diffusion Models via Discrete Denoising Posterior Prediction
Oct 10, 2024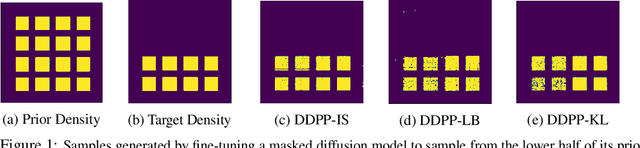
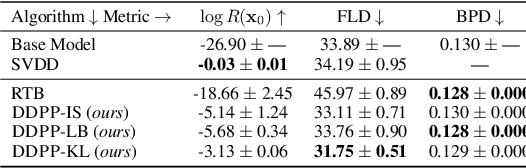
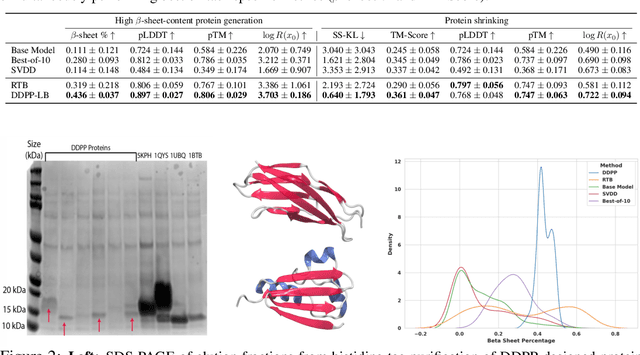
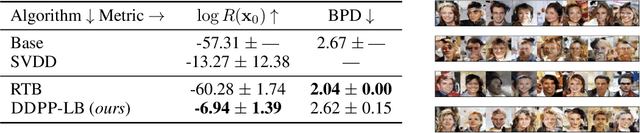
Abstract:Generative modeling of discrete data underlies important applications spanning text-based agents like ChatGPT to the design of the very building blocks of life in protein sequences. However, application domains need to exert control over the generated data by steering the generative process - typically via RLHF - to satisfy a specified property, reward, or affinity metric. In this paper, we study the problem of steering Masked Diffusion Models (MDMs), a recent class of discrete diffusion models that offer a compelling alternative to traditional autoregressive models. We introduce Discrete Denoising Posterior Prediction (DDPP), a novel framework that casts the task of steering pre-trained MDMs as a problem of probabilistic inference by learning to sample from a target Bayesian posterior. Our DDPP framework leads to a family of three novel objectives that are all simulation-free, and thus scalable while applying to general non-differentiable reward functions. Empirically, we instantiate DDPP by steering MDMs to perform class-conditional pixel-level image modeling, RLHF-based alignment of MDMs using text-based rewards, and finetuning protein language models to generate more diverse secondary structures and shorter proteins. We substantiate our designs via wet-lab validation, where we observe transient expression of reward-optimized protein sequences.
E2Efold-3D: End-to-End Deep Learning Method for accurate de novo RNA 3D Structure Prediction
Jul 04, 2022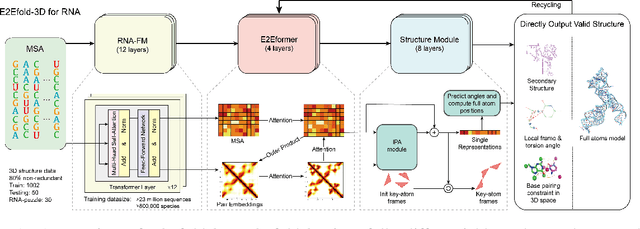
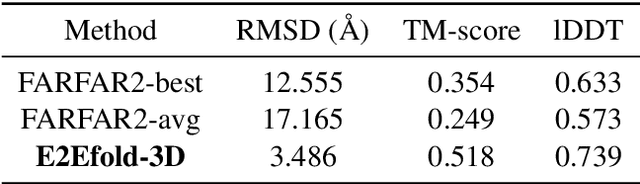
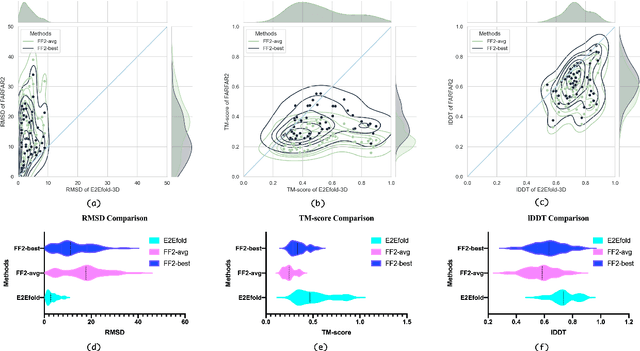
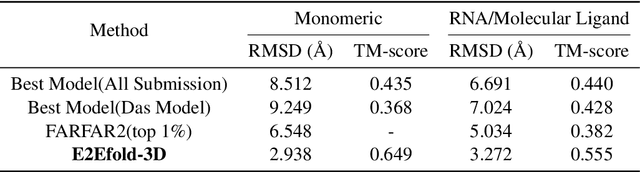
Abstract:RNA structure determination and prediction can promote RNA-targeted drug development and engineerable synthetic elements design. But due to the intrinsic structural flexibility of RNAs, all the three mainstream structure determination methods (X-ray crystallography, NMR, and Cryo-EM) encounter challenges when resolving the RNA structures, which leads to the scarcity of the resolved RNA structures. Computational prediction approaches emerge as complementary to the experimental techniques. However, none of the \textit{de novo} approaches is based on deep learning since too few structures are available. Instead, most of them apply the time-consuming sampling-based strategies, and their performance seems to hit the plateau. In this work, we develop the first end-to-end deep learning approach, E2Efold-3D, to accurately perform the \textit{de novo} RNA structure prediction. Several novel components are proposed to overcome the data scarcity, such as a fully-differentiable end-to-end pipeline, secondary structure-assisted self-distillation, and parameter-efficient backbone formulation. Such designs are validated on the independent, non-overlapping RNA puzzle testing dataset and reach an average sub-4 \AA{} root-mean-square deviation, demonstrating its superior performance compared to state-of-the-art approaches. Interestingly, it also achieves promising results when predicting RNA complex structures, a feat that none of the previous systems could accomplish. When E2Efold-3D is coupled with the experimental techniques, the RNA structure prediction field can be greatly advanced.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge