Lazar Atanackovic
Curly Flow Matching for Learning Non-gradient Field Dynamics
Oct 30, 2025Abstract:Modeling the transport dynamics of natural processes from population-level observations is a ubiquitous problem in the natural sciences. Such models rely on key assumptions about the underlying process in order to enable faithful learning of governing dynamics that mimic the actual system behavior. The de facto assumption in current approaches relies on the principle of least action that results in gradient field dynamics and leads to trajectories minimizing an energy functional between two probability measures. However, many real-world systems, such as cell cycles in single-cell RNA, are known to exhibit non-gradient, periodic behavior, which fundamentally cannot be captured by current state-of-the-art methods such as flow and bridge matching. In this paper, we introduce Curly Flow Matching (Curly-FM), a novel approach that is capable of learning non-gradient field dynamics by designing and solving a Schr\"odinger bridge problem with a non-zero drift reference process -- in stark contrast to typical zero-drift reference processes -- which is constructed using inferred velocities in addition to population snapshot data. We showcase Curly-FM by solving the trajectory inference problems for single cells, computational fluid dynamics, and ocean currents with approximate velocities. We demonstrate that Curly-FM can learn trajectories that better match both the reference process and population marginals. Curly-FM expands flow matching models beyond the modeling of populations and towards the modeling of known periodic behavior in physical systems. Our code repository is accessible at: https://github.com/kpetrovicc/curly-flow-matching.git
The Superposition of Diffusion Models Using the Itô Density Estimator
Dec 23, 2024Abstract:The Cambrian explosion of easily accessible pre-trained diffusion models suggests a demand for methods that combine multiple different pre-trained diffusion models without incurring the significant computational burden of re-training a larger combined model. In this paper, we cast the problem of combining multiple pre-trained diffusion models at the generation stage under a novel proposed framework termed superposition. Theoretically, we derive superposition from rigorous first principles stemming from the celebrated continuity equation and design two novel algorithms tailor-made for combining diffusion models in SuperDiff. SuperDiff leverages a new scalable It\^o density estimator for the log likelihood of the diffusion SDE which incurs no additional overhead compared to the well-known Hutchinson's estimator needed for divergence calculations. We demonstrate that SuperDiff is scalable to large pre-trained diffusion models as superposition is performed solely through composition during inference, and also enjoys painless implementation as it combines different pre-trained vector fields through an automated re-weighting scheme. Notably, we show that SuperDiff is efficient during inference time, and mimics traditional composition operators such as the logical OR and the logical AND. We empirically demonstrate the utility of using SuperDiff for generating more diverse images on CIFAR-10, more faithful prompt conditioned image editing using Stable Diffusion, and improved unconditional de novo structure design of proteins. https://github.com/necludov/super-diffusion
Meta Flow Matching: Integrating Vector Fields on the Wasserstein Manifold
Aug 26, 2024



Abstract:Numerous biological and physical processes can be modeled as systems of interacting entities evolving continuously over time, e.g. the dynamics of communicating cells or physical particles. Learning the dynamics of such systems is essential for predicting the temporal evolution of populations across novel samples and unseen environments. Flow-based models allow for learning these dynamics at the population level - they model the evolution of the entire distribution of samples. However, current flow-based models are limited to a single initial population and a set of predefined conditions which describe different dynamics. We argue that multiple processes in natural sciences have to be represented as vector fields on the Wasserstein manifold of probability densities. That is, the change of the population at any moment in time depends on the population itself due to the interactions between samples. In particular, this is crucial for personalized medicine where the development of diseases and their respective treatment response depends on the microenvironment of cells specific to each patient. We propose Meta Flow Matching (MFM), a practical approach to integrating along these vector fields on the Wasserstein manifold by amortizing the flow model over the initial populations. Namely, we embed the population of samples using a Graph Neural Network (GNN) and use these embeddings to train a Flow Matching model. This gives MFM the ability to generalize over the initial distributions unlike previously proposed methods. We demonstrate the ability of MFM to improve prediction of individual treatment responses on a large scale multi-patient single-cell drug screen dataset.
Investigating Generalization Behaviours of Generative Flow Networks
Feb 07, 2024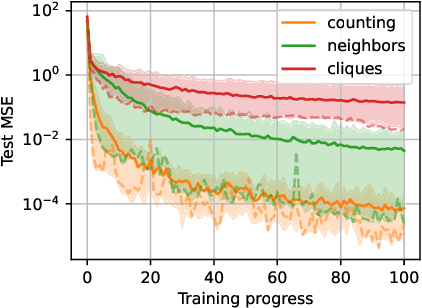

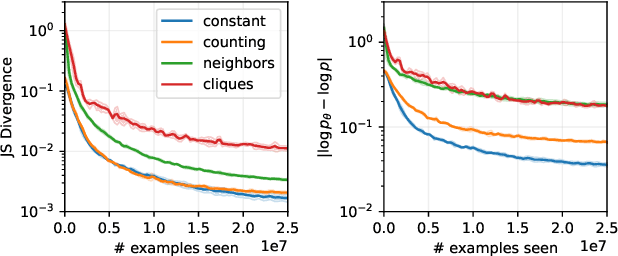

Abstract:Generative Flow Networks (GFlowNets, GFNs) are a generative framework for learning unnormalized probability mass functions over discrete spaces. Since their inception, GFlowNets have proven to be useful for learning generative models in applications where the majority of the discrete space is unvisited during training. This has inspired some to hypothesize that GFlowNets, when paired with deep neural networks (DNNs), have favourable generalization properties. In this work, we empirically verify some of the hypothesized mechanisms of generalization of GFlowNets. In particular, we find that the functions that GFlowNets learn to approximate have an implicit underlying structure which facilitate generalization. We also find that GFlowNets are sensitive to being trained offline and off-policy; however, the reward implicitly learned by GFlowNets is robust to changes in the training distribution.
A Computational Framework for Solving Wasserstein Lagrangian Flows
Oct 17, 2023Abstract:The dynamical formulation of the optimal transport can be extended through various choices of the underlying geometry ($\textit{kinetic energy}$), and the regularization of density paths ($\textit{potential energy}$). These combinations yield different variational problems ($\textit{Lagrangians}$), encompassing many variations of the optimal transport problem such as the Schr\"odinger bridge, unbalanced optimal transport, and optimal transport with physical constraints, among others. In general, the optimal density path is unknown, and solving these variational problems can be computationally challenging. Leveraging the dual formulation of the Lagrangians, we propose a novel deep learning based framework approaching all of these problems from a unified perspective. Our method does not require simulating or backpropagating through the trajectories of the learned dynamics, and does not need access to optimal couplings. We showcase the versatility of the proposed framework by outperforming previous approaches for the single-cell trajectory inference, where incorporating prior knowledge into the dynamics is crucial for correct predictions.
Simulation-free Schrödinger bridges via score and flow matching
Jul 07, 2023Abstract:We present simulation-free score and flow matching ([SF]$^2$M), a simulation-free objective for inferring stochastic dynamics given unpaired source and target samples drawn from arbitrary distributions. Our method generalizes both the score-matching loss used in the training of diffusion models and the recently proposed flow matching loss used in the training of continuous normalizing flows. [SF]$^2$M interprets continuous-time stochastic generative modeling as a Schr\"odinger bridge (SB) problem. It relies on static entropy-regularized optimal transport, or a minibatch approximation, to efficiently learn the SB without simulating the learned stochastic process. We find that [SF]$^2$M is more efficient and gives more accurate solutions to the SB problem than simulation-based methods from prior work. Finally, we apply [SF]$^2$M to the problem of learning cell dynamics from snapshot data. Notably, [SF]$^2$M is the first method to accurately model cell dynamics in high dimensions and can recover known gene regulatory networks from simulated data.
DynGFN: Bayesian Dynamic Causal Discovery using Generative Flow Networks
Feb 08, 2023



Abstract:Learning the causal structure of observable variables is a central focus for scientific discovery. Bayesian causal discovery methods tackle this problem by learning a posterior over the set of admissible graphs given our priors and observations. Existing methods primarily consider observations from static systems and assume the underlying causal structure takes the form of a directed acyclic graph (DAG). In settings with dynamic feedback mechanisms that regulate the trajectories of individual variables, this acyclicity assumption fails unless we account for time. We focus on learning Bayesian posteriors over cyclic graphs and treat causal discovery as a problem of sparse identification of a dynamical system. This imposes a natural temporal causal order between variables and captures cyclic feedback loops through time. Under this lens, we propose a new framework for Bayesian causal discovery for dynamical systems and present a novel generative flow network architecture (DynGFN) tailored for this task. Our results indicate that DynGFN learns posteriors that better encapsulate the distributions over admissible cyclic causal structures compared to counterpart state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge