Emmanuel Bengio
Torsional-GFN: a conditional conformation generator for small molecules
Jul 15, 2025Abstract:Generating stable molecular conformations is crucial in several drug discovery applications, such as estimating the binding affinity of a molecule to a target. Recently, generative machine learning methods have emerged as a promising, more efficient method than molecular dynamics for sampling of conformations from the Boltzmann distribution. In this paper, we introduce Torsional-GFN, a conditional GFlowNet specifically designed to sample conformations of molecules proportionally to their Boltzmann distribution, using only a reward function as training signal. Conditioned on a molecular graph and its local structure (bond lengths and angles), Torsional-GFN samples rotations of its torsion angles. Our results demonstrate that Torsional-GFN is able to sample conformations approximately proportional to the Boltzmann distribution for multiple molecules with a single model, and allows for zero-shot generalization to unseen bond lengths and angles coming from the MD simulations for such molecules. Our work presents a promising avenue for scaling the proposed approach to larger molecular systems, achieving zero-shot generalization to unseen molecules, and including the generation of the local structure into the GFlowNet model.
Virtual Cells: Predict, Explain, Discover
May 20, 2025Abstract:Drug discovery is fundamentally a process of inferring the effects of treatments on patients, and would therefore benefit immensely from computational models that can reliably simulate patient responses, enabling researchers to generate and test large numbers of therapeutic hypotheses safely and economically before initiating costly clinical trials. Even a more specific model that predicts the functional response of cells to a wide range of perturbations would be tremendously valuable for discovering safe and effective treatments that successfully translate to the clinic. Creating such virtual cells has long been a goal of the computational research community that unfortunately remains unachieved given the daunting complexity and scale of cellular biology. Nevertheless, recent advances in AI, computing power, lab automation, and high-throughput cellular profiling provide new opportunities for reaching this goal. In this perspective, we present a vision for developing and evaluating virtual cells that builds on our experience at Recursion. We argue that in order to be a useful tool to discover novel biology, virtual cells must accurately predict the functional response of a cell to perturbations and explain how the predicted response is a consequence of modifications to key biomolecular interactions. We then introduce key principles for designing therapeutically-relevant virtual cells, describe a lab-in-the-loop approach for generating novel insights with them, and advocate for biologically-grounded benchmarks to guide virtual cell development. Finally, we make the case that our approach to virtual cells provides a useful framework for building other models at higher levels of organization, including virtual patients. We hope that these directions prove useful to the research community in developing virtual models optimized for positive impact on drug discovery outcomes.
Solving Bayesian inverse problems with diffusion priors and off-policy RL
Mar 12, 2025Abstract:This paper presents a practical application of Relative Trajectory Balance (RTB), a recently introduced off-policy reinforcement learning (RL) objective that can asymptotically solve Bayesian inverse problems optimally. We extend the original work by using RTB to train conditional diffusion model posteriors from pretrained unconditional priors for challenging linear and non-linear inverse problems in vision, and science. We use the objective alongside techniques such as off-policy backtracking exploration to improve training. Importantly, our results show that existing training-free diffusion posterior methods struggle to perform effective posterior inference in latent space due to inherent biases.
Pretraining Generative Flow Networks with Inexpensive Rewards for Molecular Graph Generation
Mar 08, 2025



Abstract:Generative Flow Networks (GFlowNets) have recently emerged as a suitable framework for generating diverse and high-quality molecular structures by learning from rewards treated as unnormalized distributions. Previous works in this framework often restrict exploration by using predefined molecular fragments as building blocks, limiting the chemical space that can be accessed. In this work, we introduce Atomic GFlowNets (A-GFNs), a foundational generative model leveraging individual atoms as building blocks to explore drug-like chemical space more comprehensively. We propose an unsupervised pre-training approach using drug-like molecule datasets, which teaches A-GFNs about inexpensive yet informative molecular descriptors such as drug-likeliness, topological polar surface area, and synthetic accessibility scores. These properties serve as proxy rewards, guiding A-GFNs towards regions of chemical space that exhibit desirable pharmacological properties. We further implement a goal-conditioned finetuning process, which adapts A-GFNs to optimize for specific target properties. In this work, we pretrain A-GFN on a subset of ZINC dataset, and by employing robust evaluation metrics we show the effectiveness of our approach when compared to other relevant baseline methods for a wide range of drug design tasks.
Efficient Biological Data Acquisition through Inference Set Design
Oct 25, 2024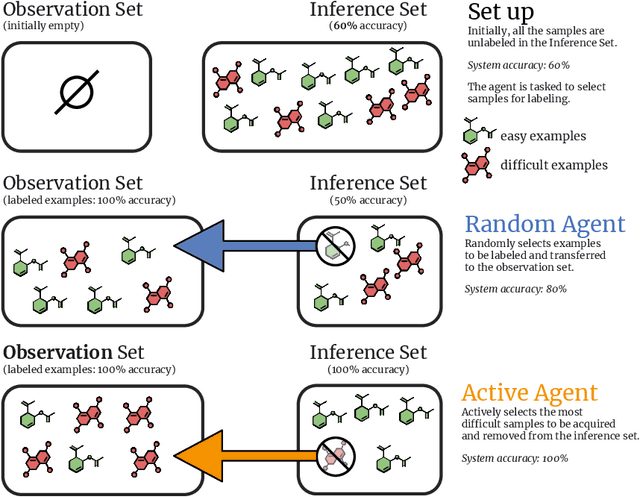

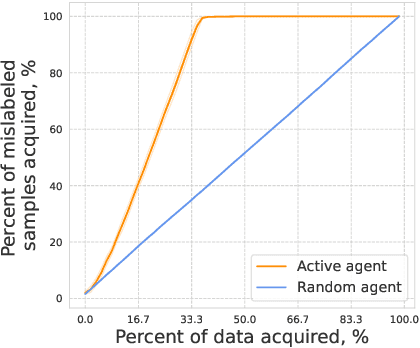

Abstract:In drug discovery, highly automated high-throughput laboratories are used to screen a large number of compounds in search of effective drugs. These experiments are expensive, so we might hope to reduce their cost by experimenting on a subset of the compounds, and predicting the outcomes of the remaining experiments. In this work, we model this scenario as a sequential subset selection problem: we aim to select the smallest set of candidates in order to achieve some desired level of accuracy for the system as a whole. Our key observation is that, if there is heterogeneity in the difficulty of the prediction problem across the input space, selectively obtaining the labels for the hardest examples in the acquisition pool will leave only the relatively easy examples to remain in the inference set, leading to better overall system performance. We call this mechanism inference set design, and propose the use of an uncertainty-based active learning solution to prune out these challenging examples. Our algorithm includes an explicit stopping criterion that stops running the experiments when it is sufficiently confident that the system has reached the target performance. Our empirical studies on image and molecular datasets, as well as a real-world large-scale biological assay, show that deploying active learning for inference set design leads to significant reduction in experimental cost while obtaining high system performance.
Action abstractions for amortized sampling
Oct 19, 2024Abstract:As trajectories sampled by policies used by reinforcement learning (RL) and generative flow networks (GFlowNets) grow longer, credit assignment and exploration become more challenging, and the long planning horizon hinders mode discovery and generalization. The challenge is particularly pronounced in entropy-seeking RL methods, such as generative flow networks, where the agent must learn to sample from a structured distribution and discover multiple high-reward states, each of which take many steps to reach. To tackle this challenge, we propose an approach to incorporate the discovery of action abstractions, or high-level actions, into the policy optimization process. Our approach involves iteratively extracting action subsequences commonly used across many high-reward trajectories and `chunking' them into a single action that is added to the action space. In empirical evaluation on synthetic and real-world environments, our approach demonstrates improved sample efficiency performance in discovering diverse high-reward objects, especially on harder exploration problems. We also observe that the abstracted high-order actions are interpretable, capturing the latent structure of the reward landscape of the action space. This work provides a cognitively motivated approach to action abstraction in RL and is the first demonstration of hierarchical planning in amortized sequential sampling.
Improved Off-policy Reinforcement Learning in Biological Sequence Design
Oct 06, 2024
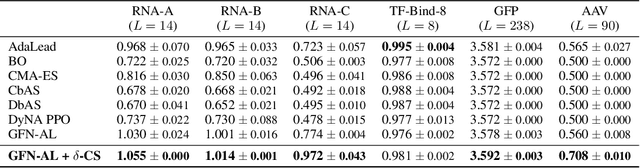
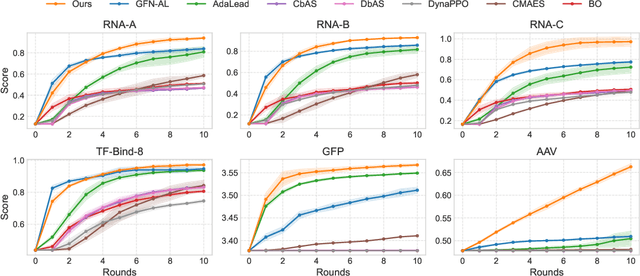
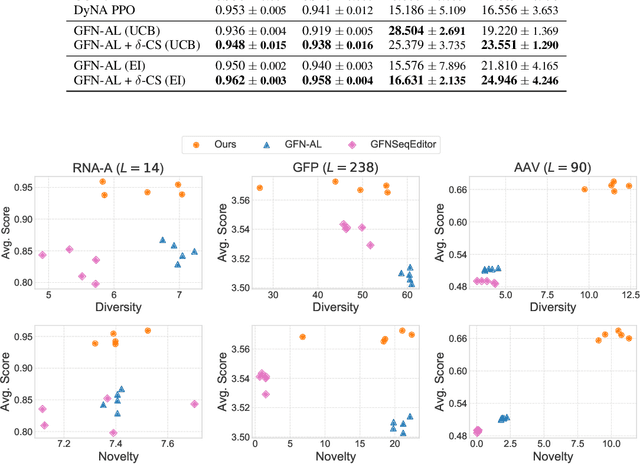
Abstract:Designing biological sequences with desired properties is a significant challenge due to the combinatorially vast search space and the high cost of evaluating each candidate sequence. To address these challenges, reinforcement learning (RL) methods, such as GFlowNets, utilize proxy models for rapid reward evaluation and annotated data for policy training. Although these approaches have shown promise in generating diverse and novel sequences, the limited training data relative to the vast search space often leads to the misspecification of proxy for out-of-distribution inputs. We introduce $\delta$-Conservative Search, a novel off-policy search method for training GFlowNets designed to improve robustness against proxy misspecification. The key idea is to incorporate conservativeness, controlled by parameter $\delta$, to constrain the search to reliable regions. Specifically, we inject noise into high-score offline sequences by randomly masking tokens with a Bernoulli distribution of parameter $\delta$ and then denoise masked tokens using the GFlowNet policy. Additionally, $\delta$ is adaptively adjusted based on the uncertainty of the proxy model for each data point. This enables the reflection of proxy uncertainty to determine the level of conservativeness. Experimental results demonstrate that our method consistently outperforms existing machine learning methods in discovering high-score sequences across diverse tasks-including DNA, RNA, protein, and peptide design-especially in large-scale scenarios.
Adaptive teachers for amortized samplers
Oct 02, 2024



Abstract:Amortized inference is the task of training a parametric model, such as a neural network, to approximate a distribution with a given unnormalized density where exact sampling is intractable. When sampling is implemented as a sequential decision-making process, reinforcement learning (RL) methods, such as generative flow networks, can be used to train the sampling policy. Off-policy RL training facilitates the discovery of diverse, high-reward candidates, but existing methods still face challenges in efficient exploration. We propose to use an adaptive training distribution (the Teacher) to guide the training of the primary amortized sampler (the Student) by prioritizing high-loss regions. The Teacher, an auxiliary behavior model, is trained to sample high-error regions of the Student and can generalize across unexplored modes, thereby enhancing mode coverage by providing an efficient training curriculum. We validate the effectiveness of this approach in a synthetic environment designed to present an exploration challenge, two diffusion-based sampling tasks, and four biochemical discovery tasks demonstrating its ability to improve sample efficiency and mode coverage.
GFlowNet Pretraining with Inexpensive Rewards
Sep 15, 2024



Abstract:Generative Flow Networks (GFlowNets), a class of generative models have recently emerged as a suitable framework for generating diverse and high-quality molecular structures by learning from unnormalized reward distributions. Previous works in this direction often restrict exploration by using predefined molecular fragments as building blocks, limiting the chemical space that can be accessed. In this work, we introduce Atomic GFlowNets (A-GFNs), a foundational generative model leveraging individual atoms as building blocks to explore drug-like chemical space more comprehensively. We propose an unsupervised pre-training approach using offline drug-like molecule datasets, which conditions A-GFNs on inexpensive yet informative molecular descriptors such as drug-likeliness, topological polar surface area, and synthetic accessibility scores. These properties serve as proxy rewards, guiding A-GFNs towards regions of chemical space that exhibit desirable pharmacological properties. We further our method by implementing a goal-conditioned fine-tuning process, which adapts A-GFNs to optimize for specific target properties. In this work, we pretrain A-GFN on the ZINC15 offline dataset and employ robust evaluation metrics to show the effectiveness of our approach when compared to other relevant baseline methods in drug design.
Baking Symmetry into GFlowNets
Jun 08, 2024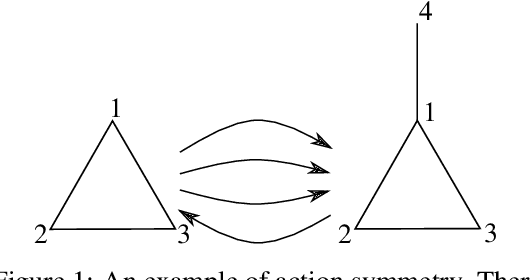

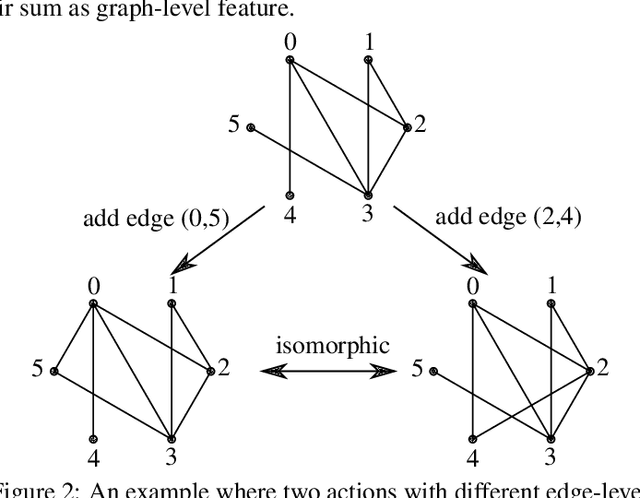

Abstract:GFlowNets have exhibited promising performance in generating diverse candidates with high rewards. These networks generate objects incrementally and aim to learn a policy that assigns probability of sampling objects in proportion to rewards. However, the current training pipelines of GFlowNets do not consider the presence of isomorphic actions, which are actions resulting in symmetric or isomorphic states. This lack of symmetry increases the amount of samples required for training GFlowNets and can result in inefficient and potentially incorrect flow functions. As a consequence, the reward and diversity of the generated objects decrease. In this study, our objective is to integrate symmetries into GFlowNets by identifying equivalent actions during the generation process. Experimental results using synthetic data demonstrate the promising performance of our proposed approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge