Pablo Lemos
Solving Bayesian inverse problems with diffusion priors and off-policy RL
Mar 12, 2025Abstract:This paper presents a practical application of Relative Trajectory Balance (RTB), a recently introduced off-policy reinforcement learning (RL) objective that can asymptotically solve Bayesian inverse problems optimally. We extend the original work by using RTB to train conditional diffusion model posteriors from pretrained unconditional priors for challenging linear and non-linear inverse problems in vision, and science. We use the objective alongside techniques such as off-policy backtracking exploration to improve training. Importantly, our results show that existing training-free diffusion posterior methods struggle to perform effective posterior inference in latent space due to inherent biases.
Amortizing intractable inference in diffusion models for vision, language, and control
May 31, 2024



Abstract:Diffusion models have emerged as effective distribution estimators in vision, language, and reinforcement learning, but their use as priors in downstream tasks poses an intractable posterior inference problem. This paper studies amortized sampling of the posterior over data, $\mathbf{x}\sim p^{\rm post}(\mathbf{x})\propto p(\mathbf{x})r(\mathbf{x})$, in a model that consists of a diffusion generative model prior $p(\mathbf{x})$ and a black-box constraint or likelihood function $r(\mathbf{x})$. We state and prove the asymptotic correctness of a data-free learning objective, relative trajectory balance, for training a diffusion model that samples from this posterior, a problem that existing methods solve only approximately or in restricted cases. Relative trajectory balance arises from the generative flow network perspective on diffusion models, which allows the use of deep reinforcement learning techniques to improve mode coverage. Experiments illustrate the broad potential of unbiased inference of arbitrary posteriors under diffusion priors: in vision (classifier guidance), language (infilling under a discrete diffusion LLM), and multimodal data (text-to-image generation). Beyond generative modeling, we apply relative trajectory balance to the problem of continuous control with a score-based behavior prior, achieving state-of-the-art results on benchmarks in offline reinforcement learning.
Sequence-Augmented SE(3)-Flow Matching For Conditional Protein Backbone Generation
May 30, 2024Abstract:Proteins are essential for almost all biological processes and derive their diverse functions from complex 3D structures, which are in turn determined by their amino acid sequences. In this paper, we exploit the rich biological inductive bias of amino acid sequences and introduce FoldFlow-2, a novel sequence-conditioned SE(3)-equivariant flow matching model for protein structure generation. FoldFlow-2 presents substantial new architectural features over the previous FoldFlow family of models including a protein large language model to encode sequence, a new multi-modal fusion trunk that combines structure and sequence representations, and a geometric transformer based decoder. To increase diversity and novelty of generated samples -- crucial for de-novo drug design -- we train FoldFlow-2 at scale on a new dataset that is an order of magnitude larger than PDB datasets of prior works, containing both known proteins in PDB and high-quality synthetic structures achieved through filtering. We further demonstrate the ability to align FoldFlow-2 to arbitrary rewards, e.g. increasing secondary structures diversity, by introducing a Reinforced Finetuning (ReFT) objective. We empirically observe that FoldFlow-2 outperforms previous state-of-the-art protein structure-based generative models, improving over RFDiffusion in terms of unconditional generation across all metrics including designability, diversity, and novelty across all protein lengths, as well as exhibiting generalization on the task of equilibrium conformation sampling. Finally, we demonstrate that a fine-tuned FoldFlow-2 makes progress on challenging conditional design tasks such as designing scaffolds for the VHH nanobody.
On diffusion models for amortized inference: Benchmarking and improving stochastic control and sampling
Feb 13, 2024
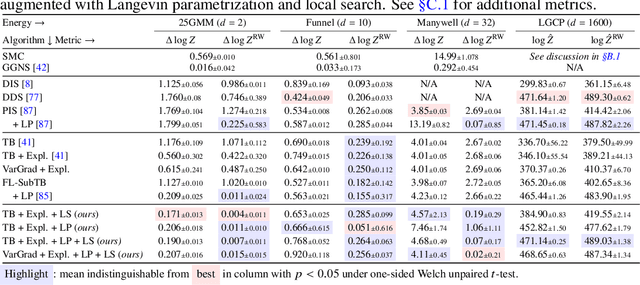
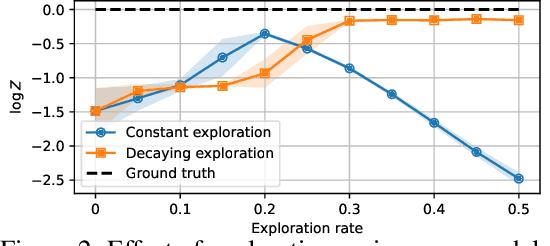
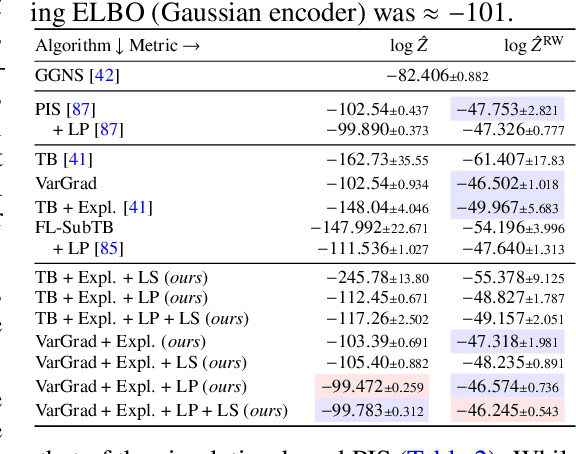
Abstract:We study the problem of training diffusion models to sample from a distribution with a given unnormalized density or energy function. We benchmark several diffusion-structured inference methods, including simulation-based variational approaches and off-policy methods (continuous generative flow networks). Our results shed light on the relative advantages of existing algorithms while bringing into question some claims from past work. We also propose a novel exploration strategy for off-policy methods, based on local search in the target space with the use of a replay buffer, and show that it improves the quality of samples on a variety of target distributions. Our code for the sampling methods and benchmarks studied is made public at https://github.com/GFNOrg/gfn-diffusion as a base for future work on diffusion models for amortized inference.
Iterated Denoising Energy Matching for Sampling from Boltzmann Densities
Feb 09, 2024



Abstract:Efficiently generating statistically independent samples from an unnormalized probability distribution, such as equilibrium samples of many-body systems, is a foundational problem in science. In this paper, we propose Iterated Denoising Energy Matching (iDEM), an iterative algorithm that uses a novel stochastic score matching objective leveraging solely the energy function and its gradient -- and no data samples -- to train a diffusion-based sampler. Specifically, iDEM alternates between (I) sampling regions of high model density from a diffusion-based sampler and (II) using these samples in our stochastic matching objective to further improve the sampler. iDEM is scalable to high dimensions as the inner matching objective, is simulation-free, and requires no MCMC samples. Moreover, by leveraging the fast mode mixing behavior of diffusion, iDEM smooths out the energy landscape enabling efficient exploration and learning of an amortized sampler. We evaluate iDEM on a suite of tasks ranging from standard synthetic energy functions to invariant $n$-body particle systems. We show that the proposed approach achieves state-of-the-art performance on all metrics and trains $2-5\times$ faster, which allows it to be the first method to train using energy on the challenging $55$-particle Lennard-Jones system.
PQMass: Probabilistic Assessment of the Quality of Generative Models using Probability Mass Estimation
Feb 06, 2024



Abstract:We propose a comprehensive sample-based method for assessing the quality of generative models. The proposed approach enables the estimation of the probability that two sets of samples are drawn from the same distribution, providing a statistically rigorous method for assessing the performance of a single generative model or the comparison of multiple competing models trained on the same dataset. This comparison can be conducted by dividing the space into non-overlapping regions and comparing the number of data samples in each region. The method only requires samples from the generative model and the test data. It is capable of functioning directly on high-dimensional data, obviating the need for dimensionality reduction. Significantly, the proposed method does not depend on assumptions regarding the density of the true distribution, and it does not rely on training or fitting any auxiliary models. Instead, it focuses on approximating the integral of the density (probability mass) across various sub-regions within the data space.
LtU-ILI: An All-in-One Framework for Implicit Inference in Astrophysics and Cosmology
Feb 06, 2024



Abstract:This paper presents the Learning the Universe Implicit Likelihood Inference (LtU-ILI) pipeline, a codebase for rapid, user-friendly, and cutting-edge machine learning (ML) inference in astrophysics and cosmology. The pipeline includes software for implementing various neural architectures, training schema, priors, and density estimators in a manner easily adaptable to any research workflow. It includes comprehensive validation metrics to assess posterior estimate coverage, enhancing the reliability of inferred results. Additionally, the pipeline is easily parallelizable, designed for efficient exploration of modeling hyperparameters. To demonstrate its capabilities, we present real applications across a range of astrophysics and cosmology problems, such as: estimating galaxy cluster masses from X-ray photometry; inferring cosmology from matter power spectra and halo point clouds; characterising progenitors in gravitational wave signals; capturing physical dust parameters from galaxy colors and luminosities; and establishing properties of semi-analytic models of galaxy formation. We also include exhaustive benchmarking and comparisons of all implemented methods as well as discussions about the challenges and pitfalls of ML inference in astronomical sciences. All code and examples are made publicly available at https://github.com/maho3/ltu-ili.
Improving Gradient-guided Nested Sampling for Posterior Inference
Dec 06, 2023Abstract:We present a performant, general-purpose gradient-guided nested sampling algorithm, ${\tt GGNS}$, combining the state of the art in differentiable programming, Hamiltonian slice sampling, clustering, mode separation, dynamic nested sampling, and parallelization. This unique combination allows ${\tt GGNS}$ to scale well with dimensionality and perform competitively on a variety of synthetic and real-world problems. We also show the potential of combining nested sampling with generative flow networks to obtain large amounts of high-quality samples from the posterior distribution. This combination leads to faster mode discovery and more accurate estimates of the partition function.
Bayesian Imaging for Radio Interferometry with Score-Based Priors
Nov 29, 2023



Abstract:The inverse imaging task in radio interferometry is a key limiting factor to retrieving Bayesian uncertainties in radio astronomy in a computationally effective manner. We use a score-based prior derived from optical images of galaxies to recover images of protoplanetary disks from the DSHARP survey. We demonstrate that our method produces plausible posterior samples despite the misspecified galaxy prior. We show that our approach produces results which are competitive with existing radio interferometry imaging algorithms.
SimBIG: Field-level Simulation-Based Inference of Galaxy Clustering
Oct 23, 2023Abstract:We present the first simulation-based inference (SBI) of cosmological parameters from field-level analysis of galaxy clustering. Standard galaxy clustering analyses rely on analyzing summary statistics, such as the power spectrum, $P_\ell$, with analytic models based on perturbation theory. Consequently, they do not fully exploit the non-linear and non-Gaussian features of the galaxy distribution. To address these limitations, we use the {\sc SimBIG} forward modelling framework to perform SBI using normalizing flows. We apply SimBIG to a subset of the BOSS CMASS galaxy sample using a convolutional neural network with stochastic weight averaging to perform massive data compression of the galaxy field. We infer constraints on $\Omega_m = 0.267^{+0.033}_{-0.029}$ and $\sigma_8=0.762^{+0.036}_{-0.035}$. While our constraints on $\Omega_m$ are in-line with standard $P_\ell$ analyses, those on $\sigma_8$ are $2.65\times$ tighter. Our analysis also provides constraints on the Hubble constant $H_0=64.5 \pm 3.8 \ {\rm km / s / Mpc}$ from galaxy clustering alone. This higher constraining power comes from additional non-Gaussian cosmological information, inaccessible with $P_\ell$. We demonstrate the robustness of our analysis by showcasing our ability to infer unbiased cosmological constraints from a series of test simulations that are constructed using different forward models than the one used in our training dataset. This work not only presents competitive cosmological constraints but also introduces novel methods for leveraging additional cosmological information in upcoming galaxy surveys like DESI, PFS, and Euclid.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge