Lucian V. Del Priore
ImageFlowNet: Forecasting Multiscale Trajectories of Disease Progression with Irregularly-Sampled Longitudinal Medical Images
Jun 20, 2024



Abstract:The forecasting of disease progression from images is a holy grail for clinical decision making. However, this task is complicated by the inherent high dimensionality, temporal sparsity and sampling irregularity in longitudinal image acquisitions. Existing methods often rely on extracting hand-crafted features and performing time-series analysis in this vector space, leading to a loss of rich spatial information within the images. To overcome these challenges, we introduce ImageFlowNet, a novel framework that learns latent-space flow fields that evolve multiscale representations in joint embedding spaces using neural ODEs and SDEs to model disease progression in the image domain. Notably, ImageFlowNet learns multiscale joint representation spaces by combining cohorts of patients together so that information can be transferred between the patient samples. The dynamics then provide plausible trajectories of progression, with the SDE providing alternative trajectories from the same starting point. We provide theoretical insights that support our formulation of ODEs, and motivate our regularizations involving high-level visual features, latent space organization, and trajectory smoothness. We then demonstrate ImageFlowNet's effectiveness through empirical evaluations on three longitudinal medical image datasets depicting progression in retinal geographic atrophy, multiple sclerosis, and glioblastoma.
CUTS: A Fully Unsupervised Framework for Medical Image Segmentation
Sep 23, 2022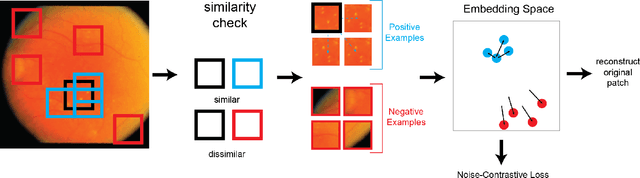
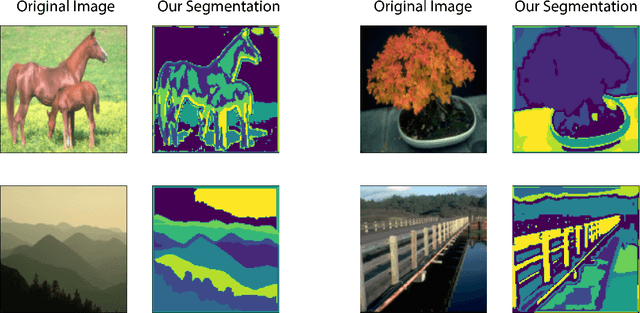
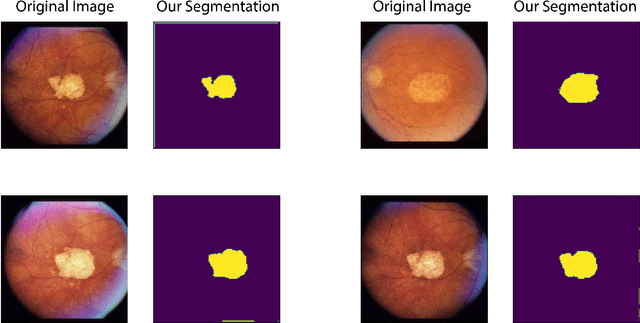
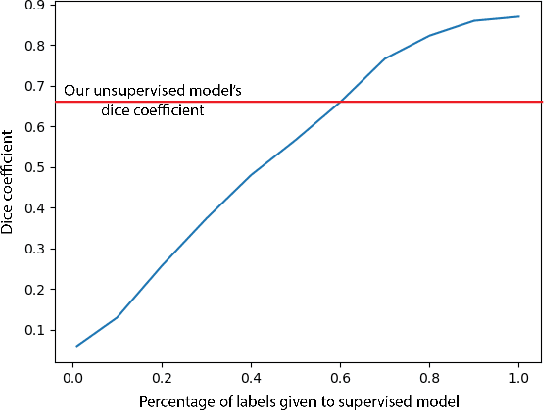
Abstract:In this work we introduce CUTS (Contrastive and Unsupervised Training for Segmentation) the first fully unsupervised deep learning framework for medical image segmentation, facilitating the use of the vast majority of imaging data that is not labeled or annotated. Segmenting medical images into regions of interest is a critical task for facilitating both patient diagnoses and quantitative research. A major limiting factor in this segmentation is the lack of labeled data, as getting expert annotations for each new set of imaging data or task can be expensive, labor intensive, and inconsistent across annotators: thus, we utilize self-supervision based on pixel-centered patches from the images themselves. Our unsupervised approach is based on a training objective with both contrastive learning and autoencoding aspects. Previous contrastive learning approaches for medical image segmentation have focused on image-level contrastive training, rather than our intra-image patch-level approach or have used this as a pre-training task where the network needed further supervised training afterwards. By contrast, we build the first entirely unsupervised framework that operates at the pixel-centered-patch level. Specifically, we add novel augmentations, a patch reconstruction loss, and introduce a new pixel clustering and identification framework. Our model achieves improved results on several key medical imaging tasks, as verified by held-out expert annotations on the task of segmenting geographic atrophy (GA) regions of images of the retina.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge