Matthew Amodio
CUTS: A Fully Unsupervised Framework for Medical Image Segmentation
Sep 23, 2022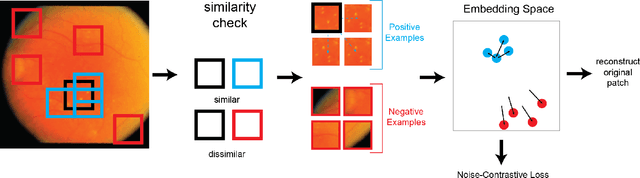
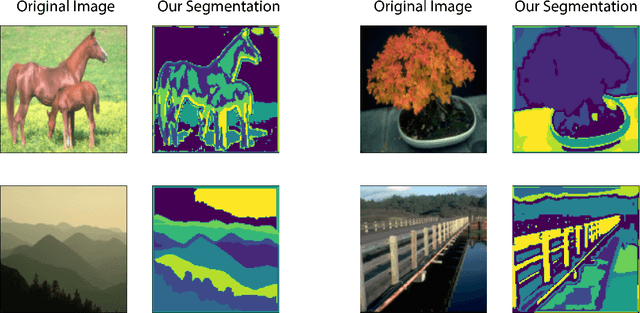
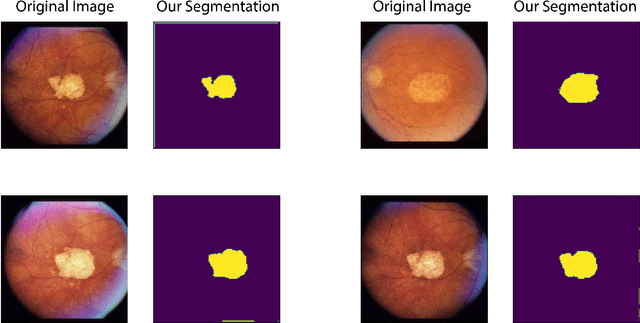
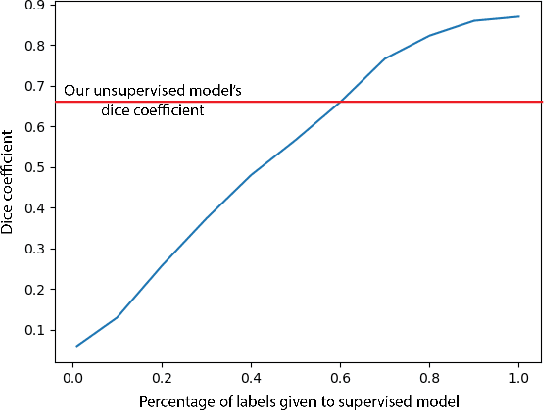
Abstract:In this work we introduce CUTS (Contrastive and Unsupervised Training for Segmentation) the first fully unsupervised deep learning framework for medical image segmentation, facilitating the use of the vast majority of imaging data that is not labeled or annotated. Segmenting medical images into regions of interest is a critical task for facilitating both patient diagnoses and quantitative research. A major limiting factor in this segmentation is the lack of labeled data, as getting expert annotations for each new set of imaging data or task can be expensive, labor intensive, and inconsistent across annotators: thus, we utilize self-supervision based on pixel-centered patches from the images themselves. Our unsupervised approach is based on a training objective with both contrastive learning and autoencoding aspects. Previous contrastive learning approaches for medical image segmentation have focused on image-level contrastive training, rather than our intra-image patch-level approach or have used this as a pre-training task where the network needed further supervised training afterwards. By contrast, we build the first entirely unsupervised framework that operates at the pixel-centered-patch level. Specifically, we add novel augmentations, a patch reconstruction loss, and introduce a new pixel clustering and identification framework. Our model achieves improved results on several key medical imaging tasks, as verified by held-out expert annotations on the task of segmenting geographic atrophy (GA) regions of images of the retina.
Image-to-image Mapping with Many Domains by Sparse Attribute Transfer
Jun 23, 2020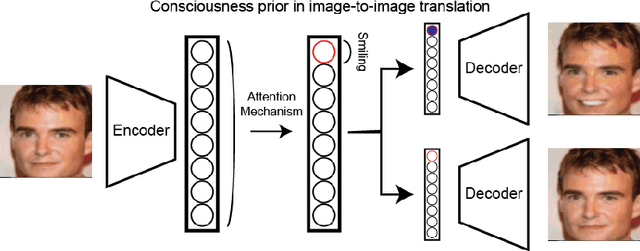
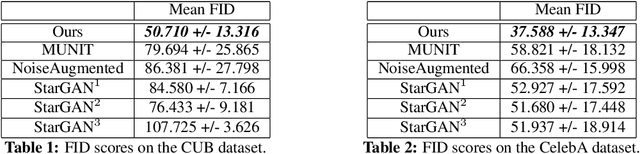
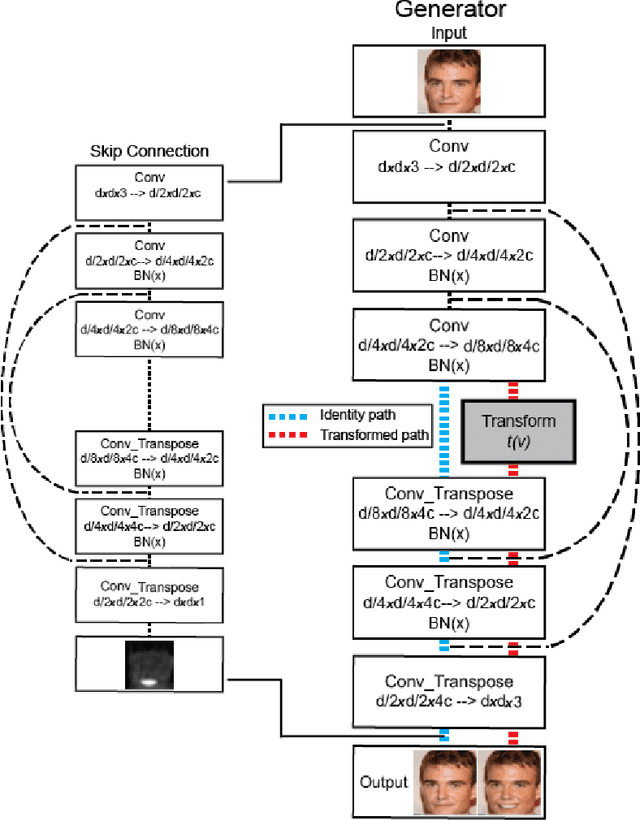
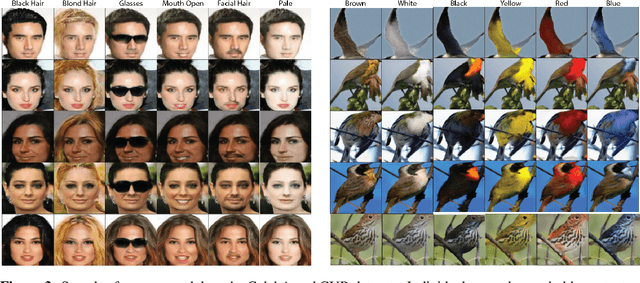
Abstract:Unsupervised image-to-image translation consists of learning a pair of mappings between two domains without known pairwise correspondences between points. The current convention is to approach this task with cycle-consistent GANs: using a discriminator to encourage the generator to change the image to match the target domain, while training the generator to be inverted with another mapping. While ending up with paired inverse functions may be a good end result, enforcing this restriction at all times during training can be a hindrance to effective modeling. We propose an alternate approach that directly restricts the generator to performing a simple sparse transformation in a latent layer, motivated by recent work from cognitive neuroscience suggesting an architectural prior on representations corresponding to consciousness. Our biologically motivated approach leads to representations more amenable to transformation by disentangling high-level abstract concepts in the latent space. We demonstrate that image-to-image domain translation with many different domains can be learned more effectively with our architecturally constrained, simple transformation than with previous unconstrained architectures that rely on a cycle-consistency loss.
TraVeLGAN: Image-to-image Translation by Transformation Vector Learning
Feb 25, 2019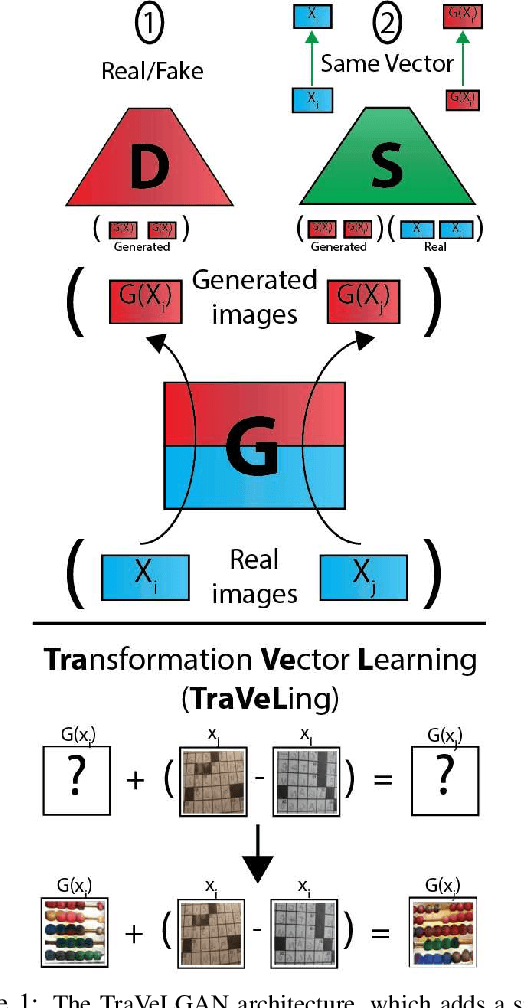
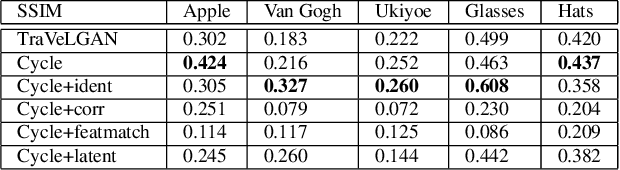
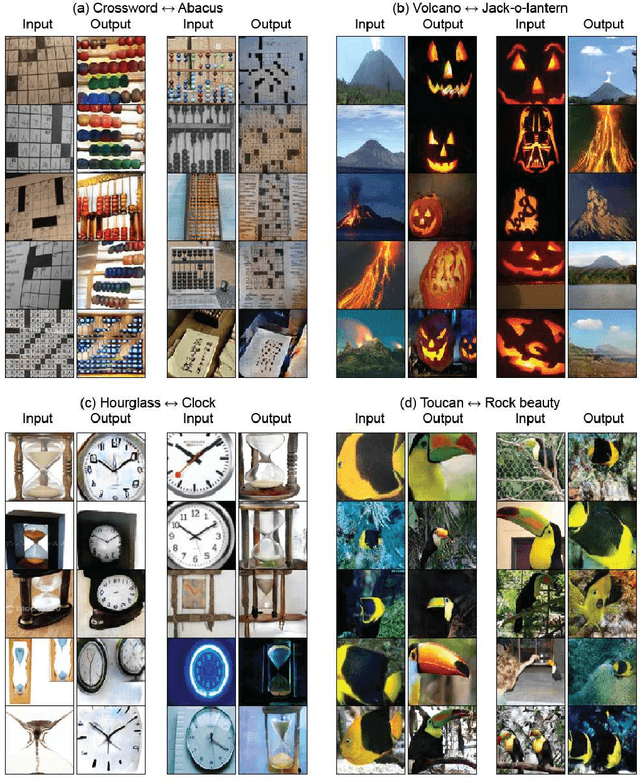
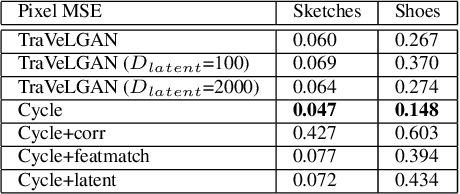
Abstract:Interest in image-to-image translation has grown substantially in recent years with the success of unsupervised models based on the cycle-consistency assumption. The achievements of these models have been limited to a particular subset of domains where this assumption yields good results, namely homogeneous domains that are characterized by style or texture differences. We tackle the challenging problem of image-to-image translation where the domains are defined by high-level shapes and contexts, as well as including significant clutter and heterogeneity. For this purpose, we introduce a novel GAN based on preserving intra-domain vector transformations in a latent space learned by a siamese network. The traditional GAN system introduced a discriminator network to guide the generator into generating images in the target domain. To this two-network system we add a third: a siamese network that guides the generator so that each original image shares semantics with its generated version. With this new three-network system, we no longer need to constrain the generators with the ubiquitous cycle-consistency restraint. As a result, the generators can learn mappings between more complex domains that differ from each other by large differences - not just style or texture.
Finding Archetypal Spaces for Data Using Neural Networks
Jan 25, 2019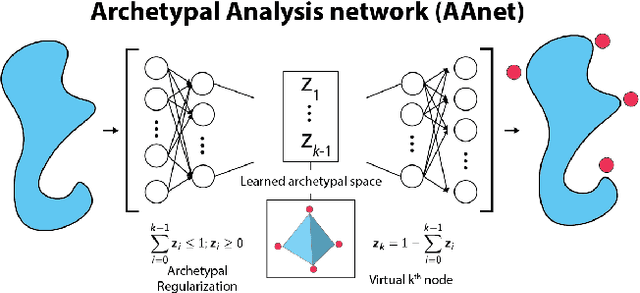
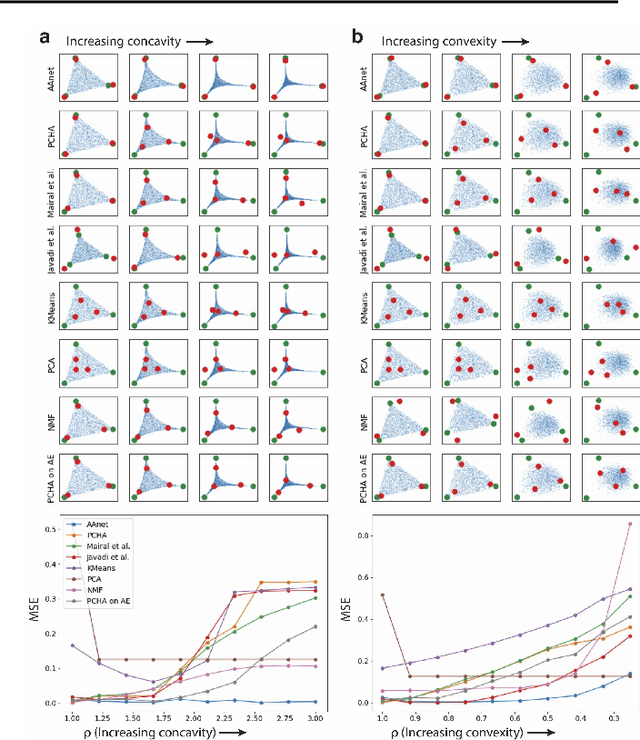
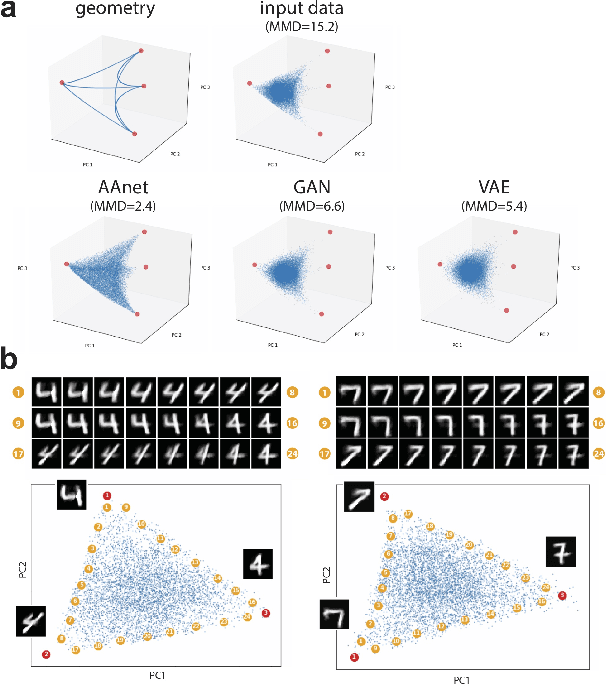
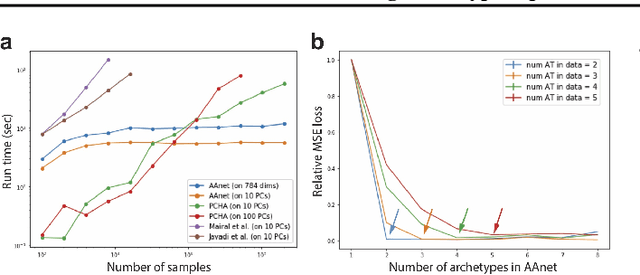
Abstract:Archetypal analysis is a type of factor analysis where data is fit by a convex polytope whose corners are "archetypes" of the data, with the data represented as a convex combination of these archetypal points. While archetypal analysis has been used on biological data, it has not achieved widespread adoption because most data are not well fit by a convex polytope in either the ambient space or after standard data transformations. We propose a new approach to archetypal analysis. Instead of fitting a convex polytope directly on data or after a specific data transformation, we train a neural network (AAnet) to learn a transformation under which the data can best fit into a polytope. We validate this approach on synthetic data where we add nonlinearity. Here, AAnet is the only method that correctly identifies the archetypes. We also demonstrate AAnet on two biological datasets. In a T cell dataset measured with single cell RNA-sequencing, AAnet identifies several archetypal states corresponding to naive, memory, and cytotoxic T cells. In a dataset of gut microbiome profiles, AAnet recovers both previously described microbiome states and identifies novel extrema in the data. Finally, we show that AAnet has generative properties allowing us to uniformly sample from the data geometry even when the input data is not uniformly distributed.
Generating and Aligning from Data Geometries with Generative Adversarial Networks
Jan 24, 2019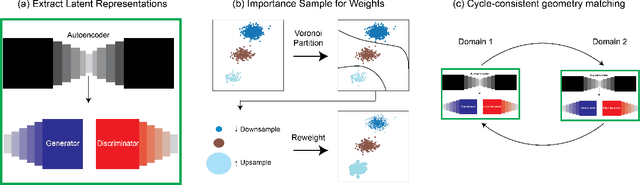
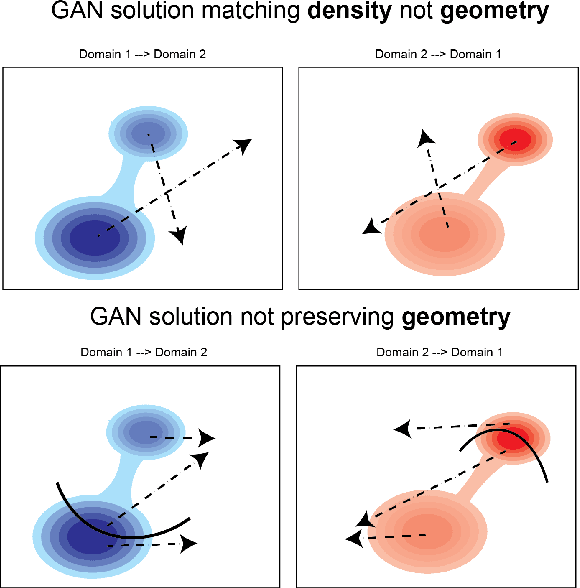
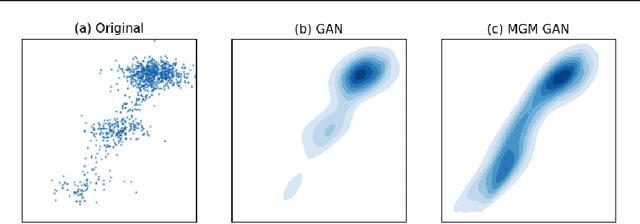
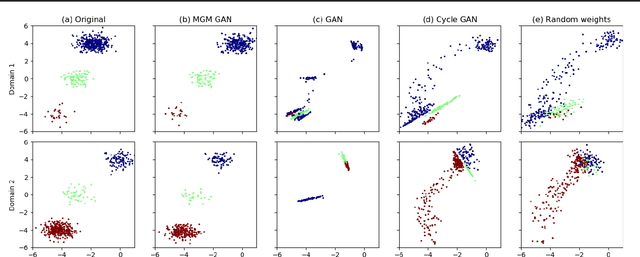
Abstract:Unsupervised domain mapping has attracted substantial attention in recent years due to the success of models based on the cycle-consistency assumption. These models map between two domains by fooling a probabilistic discriminator, thereby matching the probability distributions of the real and generated data. Instead of this probabilistic approach, we cast the problem in terms of aligning the geometry of the manifolds of the two domains. We introduce the Manifold Geometry Matching Generative Adversarial Network (MGM GAN), which adds two novel mechanisms to facilitate GANs sampling from the geometry of the manifold rather than the density and then aligning two manifold geometries: (1) an importance sampling technique that reweights points based on their density on the manifold, making the discriminator only able to discern geometry and (2) a penalty adapted from traditional manifold alignment literature that explicitly enforces the geometry to be preserved. The MGM GAN leverages the manifolds arising from a pre-trained autoencoder to bridge the gap between formal manifold alignment literature and existing GAN work, and demonstrate the advantages of modeling the manifold geometry over its density.
Graph Spectral Regularization for Neural Network Interpretability
Oct 02, 2018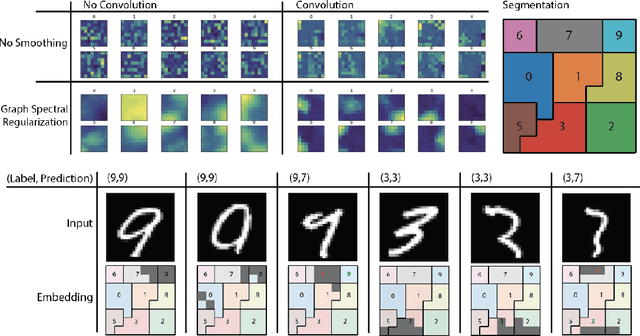
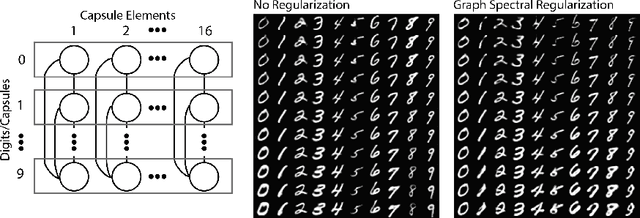
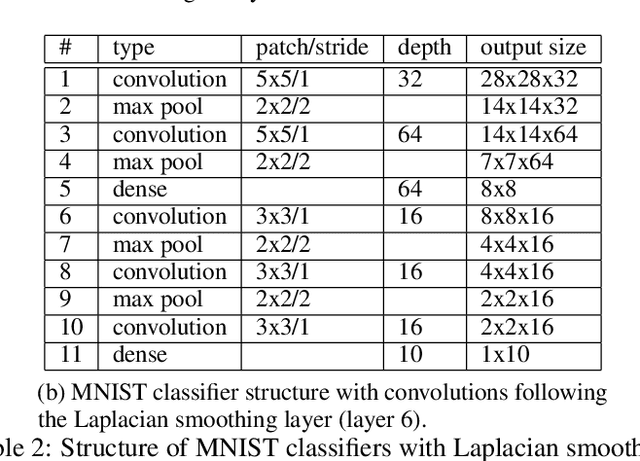
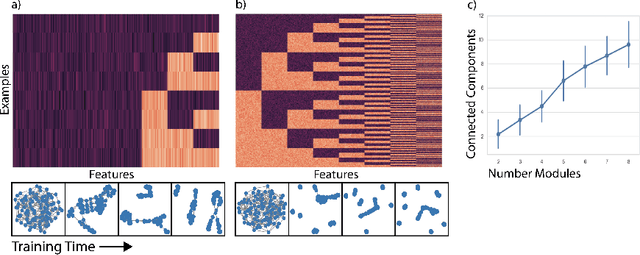
Abstract:Deep neural networks can learn meaningful representations of data. However, these representations are hard to interpret. For example, visualizing a latent layer is generally only possible for at most three dimensions. Neural networks are able to learn and benefit from much higher dimensional representationsm but these are not visually interpretable because neurons have arbitrary ordering within a layer. Here, we utilize the ability of a human observer to identify patterns in structured representations to visualize higher dimensions. To do so, we propose a class of regularizations we call Graph Spectral Regularizations that impose graph structure on latent layers. This is achieved by treating activations as signals on a predefined graph and constraining those activations using graph filters, such as low pass and wavelet-like filters. This framework allows for any kind of graphs and filters to achieve a wide range of structured regularizations depending on the inference needs of the data. First, we show a synthetic example where a graph-structured layer reveals topological features of the data. Next, we show that a smoothing regularization imposes semantically consistent ordering of nodes when applied to capsule nets. Further, we show that the graph-structured layer, using wavelet-like spatially localized filters, can form local receptive fields for improved image and biomedical data interpretation. In other words, the mapping between latent layer, neurons and the output space becomes clear due to the localization of the activations. Finally, we show that when structured as a grid, the representations create coherent images that allow for image processing techniques such as convolutions.
MAGAN: Aligning Biological Manifolds
Feb 10, 2018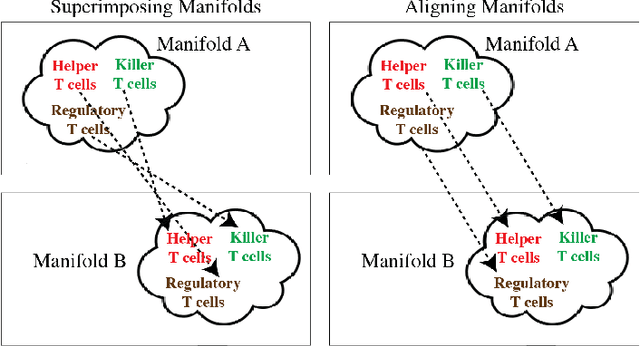
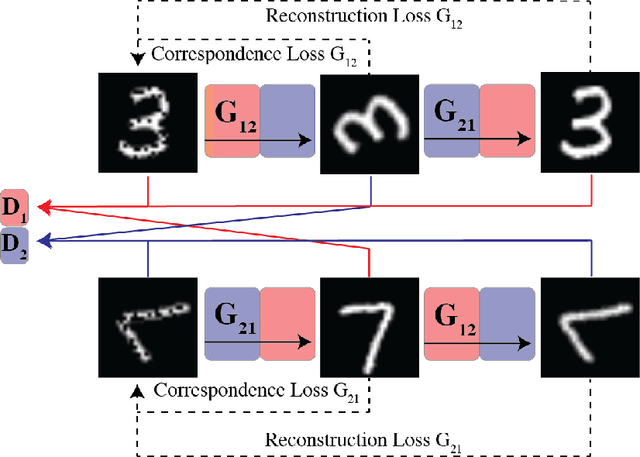
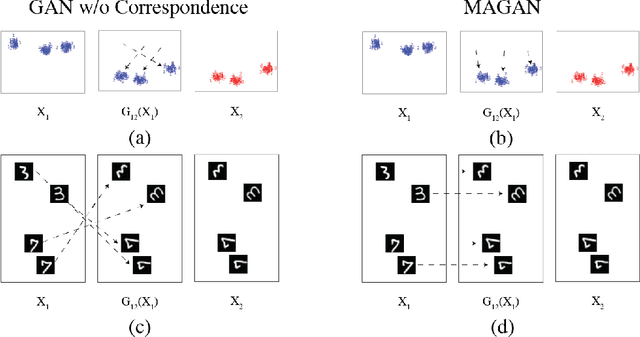
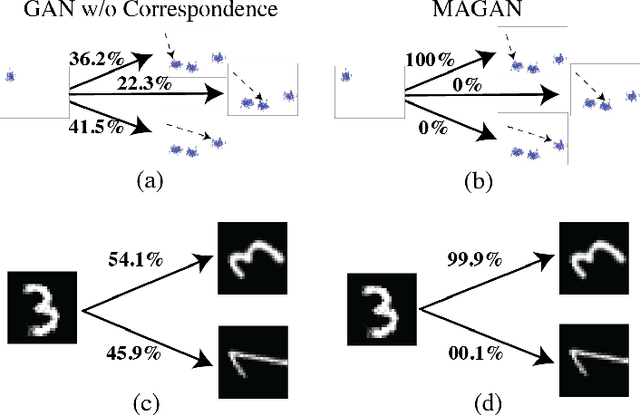
Abstract:It is increasingly common in many types of natural and physical systems (especially biological systems) to have different types of measurements performed on the same underlying system. In such settings, it is important to align the manifolds arising from each measurement in order to integrate such data and gain an improved picture of the system. We tackle this problem using generative adversarial networks (GANs). Recently, GANs have been utilized to try to find correspondences between sets of samples. However, these GANs are not explicitly designed for proper alignment of manifolds. We present a new GAN called the Manifold-Aligning GAN (MAGAN) that aligns two manifolds such that related points in each measurement space are aligned together. We demonstrate applications of MAGAN in single-cell biology in integrating two different measurement types together. In our demonstrated examples, cells from the same tissue are measured with both genomic (single-cell RNA-sequencing) and proteomic (mass cytometry) technologies. We show that the MAGAN successfully aligns them such that known correlations between measured markers are improved compared to other recently proposed models.
Neural Attribute Machines for Program Generation
May 31, 2017
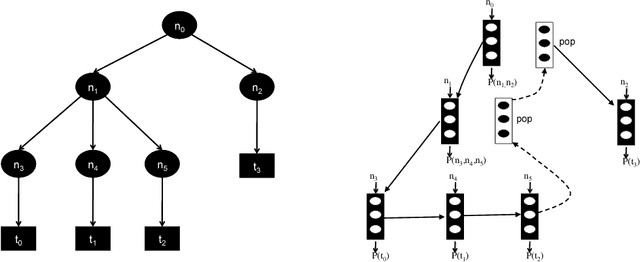
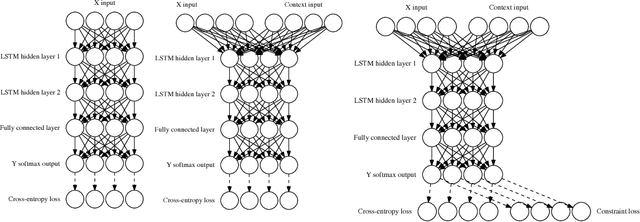
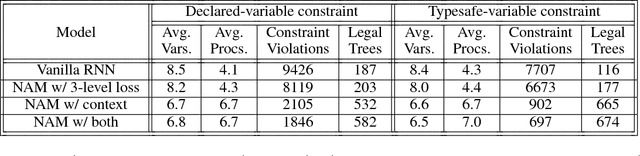
Abstract:Recurrent neural networks have achieved remarkable success at generating sequences with complex structures, thanks to advances that include richer embeddings of input and cures for vanishing gradients. Trained only on sequences from a known grammar, though, they can still struggle to learn rules and constraints of the grammar. Neural Attribute Machines (NAMs) are equipped with a logical machine that represents the underlying grammar, which is used to teach the constraints to the neural machine by (i) augmenting the input sequence, and (ii) optimizing a custom loss function. Unlike traditional RNNs, NAMs are exposed to the grammar, as well as samples from the language of the grammar. During generation, NAMs make significantly fewer violations of the constraints of the underlying grammar than RNNs trained only on samples from the language of the grammar.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge