The legibility of the imaged human brain
Paper and Code
Aug 23, 2023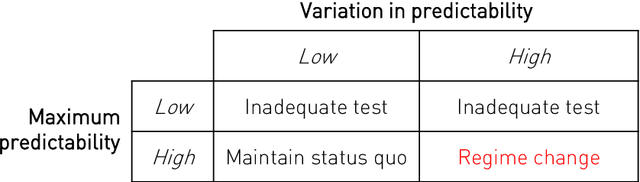

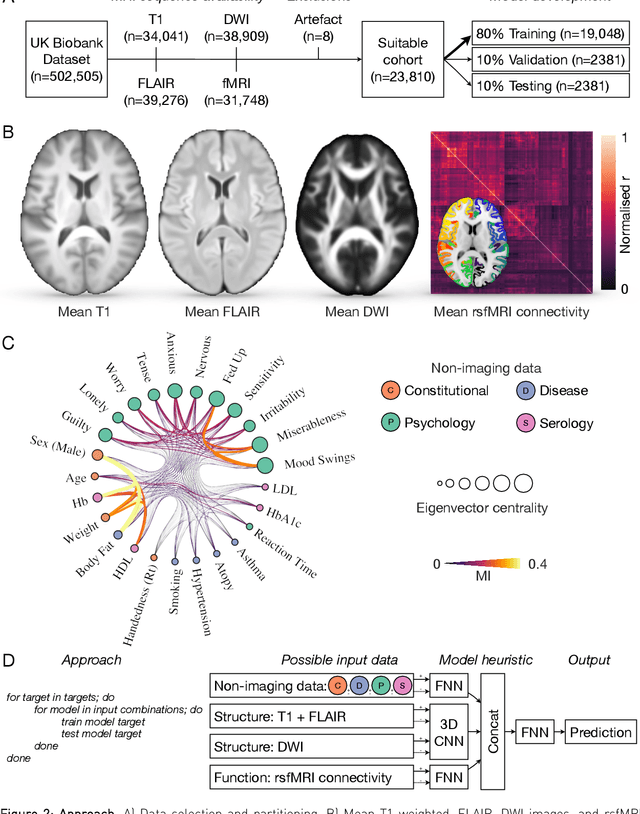
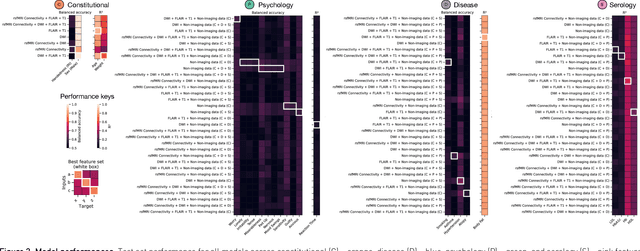
Our knowledge of the organisation of the human brain at the population-level is yet to translate into power to predict functional differences at the individual-level, limiting clinical applications, and casting doubt on the generalisability of inferred mechanisms. It remains unknown whether the difficulty arises from the absence of individuating biological patterns within the brain, or from limited power to access them with the models and compute at our disposal. Here we comprehensively investigate the resolvability of such patterns with data and compute at unprecedented scale. Across 23810 unique participants from UK Biobank, we systematically evaluate the predictability of 25 individual biological characteristics, from all available combinations of structural and functional neuroimaging data. Over 4526 GPU*hours of computation, we train, optimize, and evaluate out-of-sample 700 individual predictive models, including multilayer perceptrons of demographic, psychological, serological, chronic morbidity, and functional connectivity characteristics, and both uni- and multi-modal 3D convolutional neural network models of macro- and micro-structural brain imaging. We find a marked discrepancy between the high predictability of sex (balanced accuracy 99.7%), age (mean absolute error 2.048 years, R2 0.859), and weight (mean absolute error 2.609Kg, R2 0.625), for which we set new state-of-the-art performance, and the surprisingly low predictability of other characteristics. Neither structural nor functional imaging predicted individual psychology better than the coincidence of common chronic morbidity (p<0.05). Serology predicted common morbidity (p<0.05) and was best predicted by it (p<0.001), followed by structural neuroimaging (p<0.05). Our findings suggest either more informative imaging or more powerful models will be needed to decipher individual level characteristics from the brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge