Yaël Balbastre
Neurovascular Segmentation in sOCT with Deep Learning and Synthetic Training Data
Jul 01, 2024



Abstract:Microvascular anatomy is known to be involved in various neurological disorders. However, understanding these disorders is hindered by the lack of imaging modalities capable of capturing the comprehensive three-dimensional vascular network structure at microscopic resolution. With a lateral resolution of $<=$20 {\textmu}m and ability to reconstruct large tissue blocks up to tens of cubic centimeters, serial-section optical coherence tomography (sOCT) is well suited for this task. This method uses intrinsic optical properties to visualize the vessels and therefore does not possess a specific contrast, which complicates the extraction of accurate vascular models. The performance of traditional vessel segmentation methods is heavily degraded in the presence of substantial noise and imaging artifacts and is sensitive to domain shifts, while convolutional neural networks (CNNs) require extensive labeled data and are also sensitive the precise intensity characteristics of the data that they are trained on. Building on the emerging field of synthesis-based training, this study demonstrates a synthesis engine for neurovascular segmentation in sOCT images. Characterized by minimal priors and high variance sampling, our highly generalizable method tested on five distinct sOCT acquisitions eliminates the need for manual annotations while attaining human-level precision. Our approach comprises two phases: label synthesis and label-to-image transformation. We demonstrate the efficacy of the former by comparing it to several more realistic sets of training labels, and the latter by an ablation study of synthetic noise and artifact models.
Fully Convolutional Slice-to-Volume Reconstruction for Single-Stack MRI
Dec 05, 2023



Abstract:In magnetic resonance imaging (MRI), slice-to-volume reconstruction (SVR) refers to computational reconstruction of an unknown 3D magnetic resonance volume from stacks of 2D slices corrupted by motion. While promising, current SVR methods require multiple slice stacks for accurate 3D reconstruction, leading to long scans and limiting their use in time-sensitive applications such as fetal fMRI. Here, we propose a SVR method that overcomes the shortcomings of previous work and produces state-of-the-art reconstructions in the presence of extreme inter-slice motion. Inspired by the recent success of single-view depth estimation methods, we formulate SVR as a single-stack motion estimation task and train a fully convolutional network to predict a motion stack for a given slice stack, producing a 3D reconstruction as a byproduct of the predicted motion. Extensive experiments on the SVR of adult and fetal brains demonstrate that our fully convolutional method is twice as accurate as previous SVR methods. Our code is available at github.com/seannz/svr.
Diffeomorphic Multi-Resolution Deep Learning Registration for Applications in Breast MRI
Sep 24, 2023



Abstract:In breast surgical planning, accurate registration of MR images across patient positions has the potential to improve the localisation of tumours during breast cancer treatment. While learning-based registration methods have recently become the state-of-the-art approach for most medical image registration tasks, these methods have yet to make inroads into breast image registration due to certain difficulties-the lack of rich texture information in breast MR images and the need for the deformations to be diffeomophic. In this work, we propose learning strategies for breast MR image registration that are amenable to diffeomorphic constraints, together with early experimental results from in-silico and in-vivo experiments. One key contribution of this work is a registration network which produces superior registration outcomes for breast images in addition to providing diffeomorphic guarantees.
SuperWarp: Supervised Learning and Warping on U-Net for Invariant Subvoxel-Precise Registration
May 15, 2022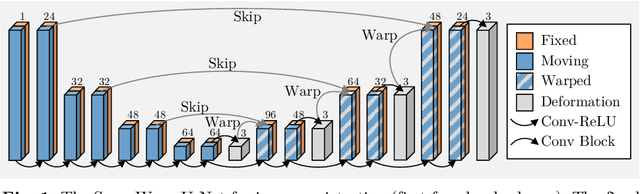

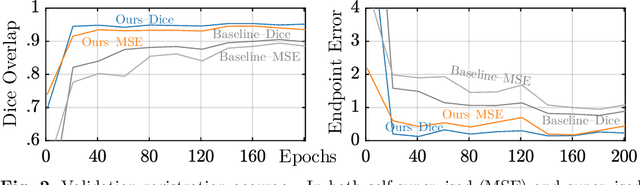
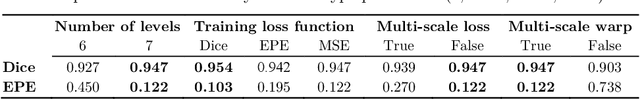
Abstract:In recent years, learning-based image registration methods have gradually moved away from direct supervision with target warps to instead use self-supervision, with excellent results in several registration benchmarks. These approaches utilize a loss function that penalizes the intensity differences between the fixed and moving images, along with a suitable regularizer on the deformation. In this paper, we argue that the relative failure of supervised registration approaches can in part be blamed on the use of regular U-Nets, which are jointly tasked with feature extraction, feature matching, and estimation of deformation. We introduce one simple but crucial modification to the U-Net that disentangles feature extraction and matching from deformation prediction, allowing the U-Net to warp the features, across levels, as the deformation field is evolved. With this modification, direct supervision using target warps begins to outperform self-supervision approaches that require segmentations, presenting new directions for registration when images do not have segmentations. We hope that our findings in this preliminary workshop paper will re-ignite research interest in supervised image registration techniques. Our code is publicly available from https://github.com/balbasty/superwarp.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021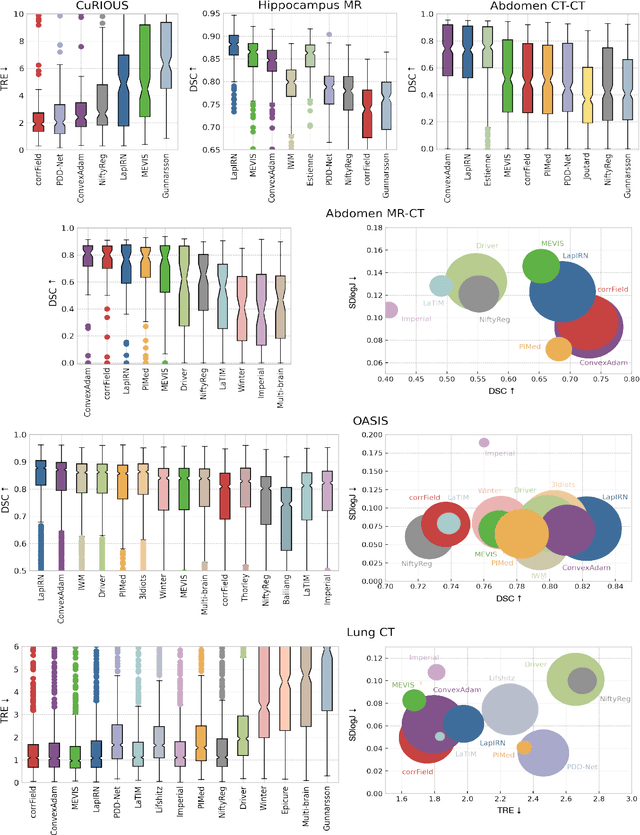
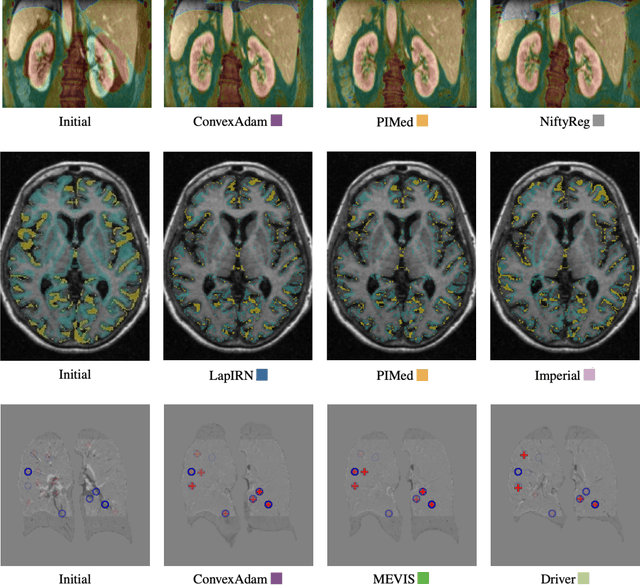
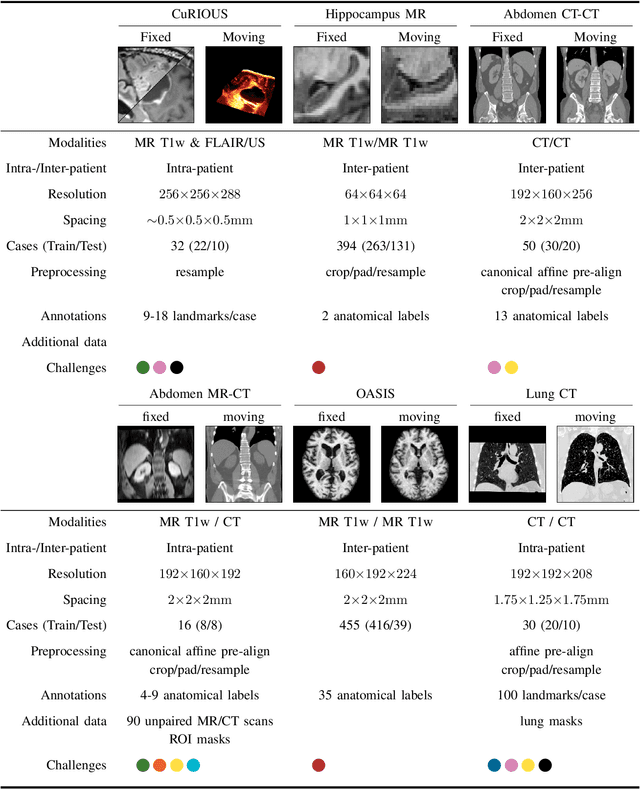
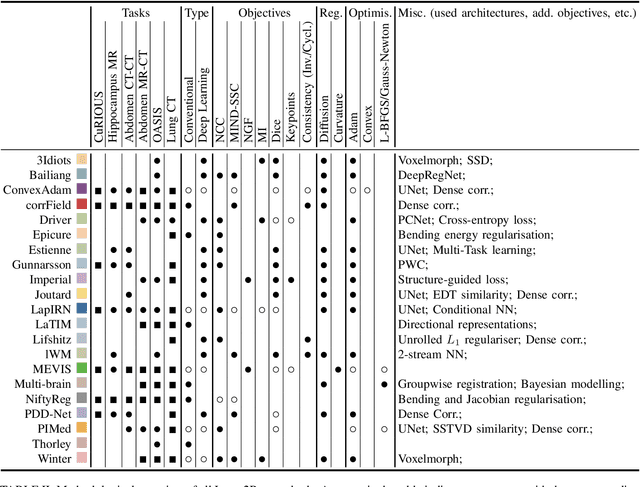
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Correcting inter-scan motion artefacts in quantitative R1 mapping at 7T
Aug 24, 2021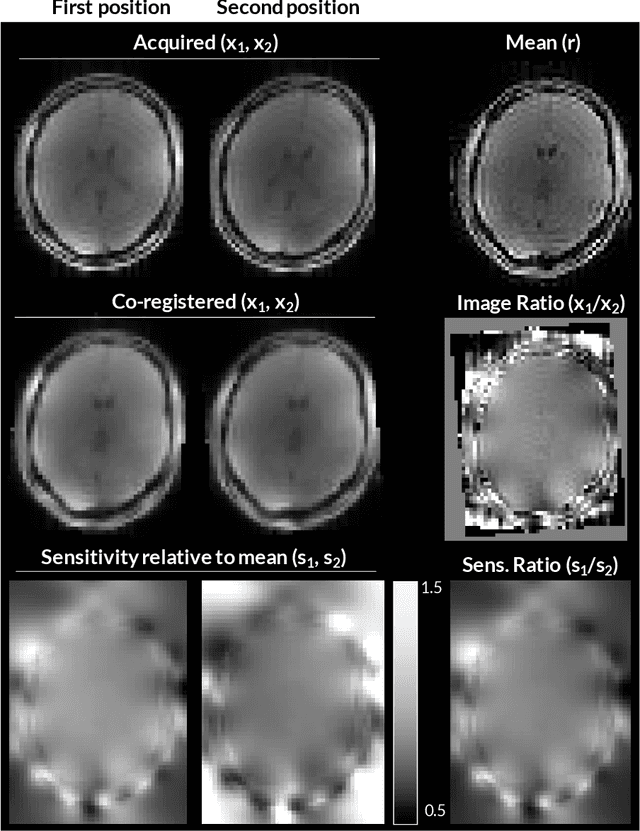
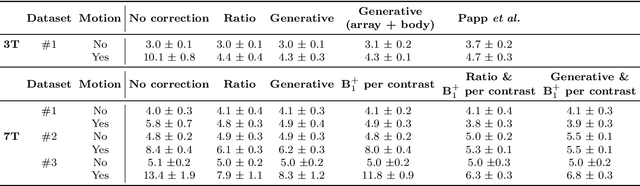
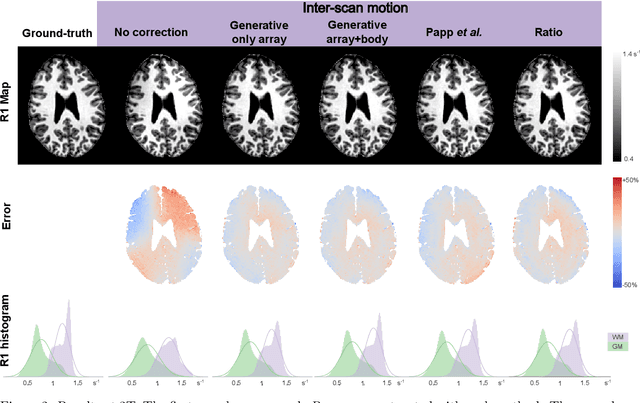
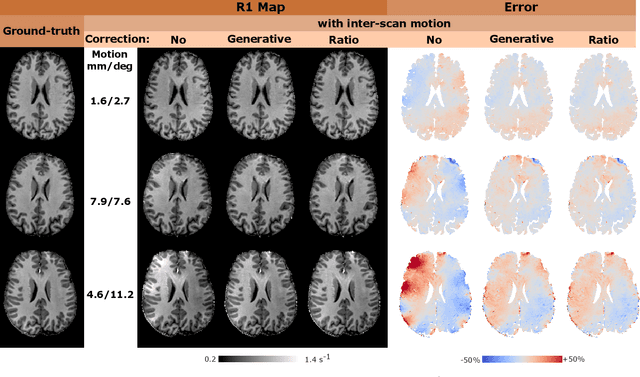
Abstract:Purpose: Inter-scan motion is a substantial source of error in $R_1$ estimation, and can be expected to increase at 7T where $B_1$ fields are more inhomogeneous. The established correction scheme does not translate to 7T since it requires a body coil reference. Here we introduce two alternatives that outperform the established method. Since they compute relative sensitivities they do not require body coil images. Theory: The proposed methods use coil-combined magnitude images to obtain the relative coil sensitivities. The first method efficiently computes the relative sensitivities via a simple ratio; the second by fitting a more sophisticated generative model. Methods: $R_1$ maps were computed using the variable flip angle (VFA) approach. Multiple datasets were acquired at 3T and 7T, with and without motion between the acquisition of the VFA volumes. $R_1$ maps were constructed without correction, with the proposed corrections, and (at 3T) with the previously established correction scheme. Results: At 3T, the proposed methods outperform the baseline method. Inter-scan motion artefacts were also reduced at 7T. However, reproducibility only converged on that of the no motion condition if position-specific transmit field effects were also incorporated. Conclusion: The proposed methods simplify inter-scan motion correction of $R_1$ maps and are applicable at both 3T and 7T, where a body coil is typically not available. The open-source code for all methods is made publicly available.
An MRF-UNet Product of Experts for Image Segmentation
Apr 12, 2021



Abstract:While convolutional neural networks (CNNs) trained by back-propagation have seen unprecedented success at semantic segmentation tasks, they are known to struggle on out-of-distribution data. Markov random fields (MRFs) on the other hand, encode simpler distributions over labels that, although less flexible than UNets, are less prone to over-fitting. In this paper, we propose to fuse both strategies by computing the product of distributions of a UNet and an MRF. As this product is intractable, we solve for an approximate distribution using an iterative mean-field approach. The resulting MRF-UNet is trained jointly by back-propagation. Compared to other works using conditional random fields (CRFs), the MRF has no dependency on the imaging data, which should allow for less over-fitting. We show on 3D neuroimaging data that this novel network improves generalisation to out-of-distribution samples. Furthermore, it allows the overall number of parameters to be reduced while preserving high accuracy. These results suggest that a classic MRF smoothness prior can allow for less over-fitting when principally integrated into a CNN model. Our implementation is available at https://github.com/balbasty/nitorch.
Flexible Bayesian Modelling for Nonlinear Image Registration
Jun 03, 2020



Abstract:We describe a diffeomorphic registration algorithm that allows groups of images to be accurately aligned to a common space, which we intend to incorporate into the SPM software. The idea is to perform inference in a probabilistic graphical model that accounts for variability in both shape and appearance. The resulting framework is general and entirely unsupervised. The model is evaluated at inter-subject registration of 3D human brain scans. Here, the main modeling assumption is that individual anatomies can be generated by deforming a latent 'average' brain. The method is agnostic to imaging modality and can be applied with no prior processing. We evaluate the algorithm using freely available, manually labelled datasets. In this validation we achieve state-of-the-art results, within reasonable runtimes, against previous state-of-the-art widely used, inter-subject registration algorithms. On the unprocessed dataset, the increase in overlap score is over 17%. These results demonstrate the benefits of using informative computational anatomy frameworks for nonlinear registration.
Joint Total Variation ESTATICS for Robust Multi-Parameter Mapping
May 28, 2020


Abstract:Quantitative magnetic resonance imaging (qMRI) derives tissue-specific parameters -- such as the apparent transverse relaxation rate R2*, the longitudinal relaxation rate R1 and the magnetisation transfer saturation -- that can be compared across sites and scanners and carry important information about the underlying microstructure. The multi-parameter mapping (MPM) protocol takes advantage of multi-echo acquisitions with variable flip angles to extract these parameters in a clinically acceptable scan time. In this context, ESTATICS performs a joint loglinear fit of multiple echo series to extract R2* and multiple extrapolated intercepts, thereby improving robustness to motion and decreasing the variance of the estimators. In this paper, we extend this model in two ways: (1) by introducing a joint total variation (JTV) prior on the intercepts and decay, and (2) by deriving a nonlinear maximum \emph{a posteriori} estimate. We evaluated the proposed algorithm by predicting left-out echoes in a rich single-subject dataset. In this validation, we outperformed other state-of-the-art methods and additionally showed that the proposed approach greatly reduces the variance of the estimated maps, without introducing bias.
Groupwise Multimodal Image Registration using Joint Total Variation
May 06, 2020



Abstract:In medical imaging it is common practice to acquire a wide range of modalities (MRI, CT, PET, etc.), to highlight different structures or pathologies. As patient movement between scans or scanning session is unavoidable, registration is often an essential step before any subsequent image analysis. In this paper, we introduce a cost function based on joint total variation for such multimodal image registration. This cost function has the advantage of enabling principled, groupwise alignment of multiple images, whilst being insensitive to strong intensity non-uniformities. We evaluate our algorithm on rigidly aligning both simulated and real 3D brain scans. This validation shows robustness to strong intensity non-uniformities and low registration errors for CT/PET to MRI alignment. Our implementation is publicly available at https://github.com/brudfors/coregistration-njtv.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge