Ying Hu
Training Together, Diagnosing Better: Federated Learning for Collagen VI-Related Dystrophies
Dec 18, 2025Abstract:The application of Machine Learning (ML) to the diagnosis of rare diseases, such as collagen VI-related dystrophies (COL6-RD), is fundamentally limited by the scarcity and fragmentation of available data. Attempts to expand sampling across hospitals, institutions, or countries with differing regulations face severe privacy, regulatory, and logistical obstacles that are often difficult to overcome. The Federated Learning (FL) provides a promising solution by enabling collaborative model training across decentralized datasets while keeping patient data local and private. Here, we report a novel global FL initiative using the Sherpa.ai FL platform, which leverages FL across distributed datasets in two international organizations for the diagnosis of COL6-RD, using collagen VI immunofluorescence microscopy images from patient-derived fibroblast cultures. Our solution resulted in an ML model capable of classifying collagen VI patient images into the three primary pathogenic mechanism groups associated with COL6-RD: exon skipping, glycine substitution, and pseudoexon insertion. This new approach achieved an F1-score of 0.82, outperforming single-organization models (0.57-0.75). These results demonstrate that FL substantially improves diagnostic utility and generalizability compared to isolated institutional models. Beyond enabling more accurate diagnosis, we anticipate that this approach will support the interpretation of variants of uncertain significance and guide the prioritization of sequencing strategies to identify novel pathogenic variants.
Ultrasound Report Generation with Multimodal Large Language Models for Standardized Texts
May 13, 2025Abstract:Ultrasound (US) report generation is a challenging task due to the variability of US images, operator dependence, and the need for standardized text. Unlike X-ray and CT, US imaging lacks consistent datasets, making automation difficult. In this study, we propose a unified framework for multi-organ and multilingual US report generation, integrating fragment-based multilingual training and leveraging the standardized nature of US reports. By aligning modular text fragments with diverse imaging data and curating a bilingual English-Chinese dataset, the method achieves consistent and clinically accurate text generation across organ sites and languages. Fine-tuning with selective unfreezing of the vision transformer (ViT) further improves text-image alignment. Compared to the previous state-of-the-art KMVE method, our approach achieves relative gains of about 2\% in BLEU scores, approximately 3\% in ROUGE-L, and about 15\% in CIDEr, while significantly reducing errors such as missing or incorrect content. By unifying multi-organ and multi-language report generation into a single, scalable framework, this work demonstrates strong potential for real-world clinical workflows.
BitNet b1.58 2B4T Technical Report
Apr 16, 2025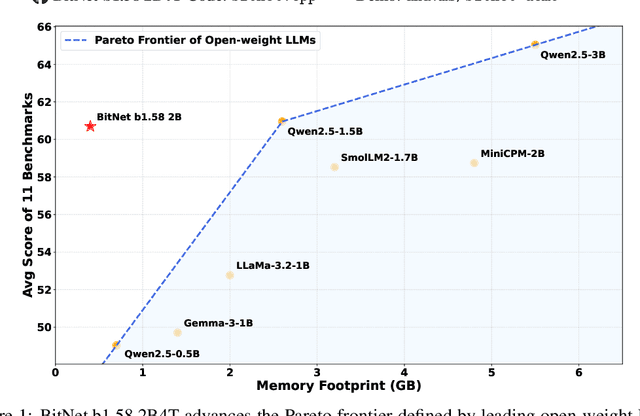
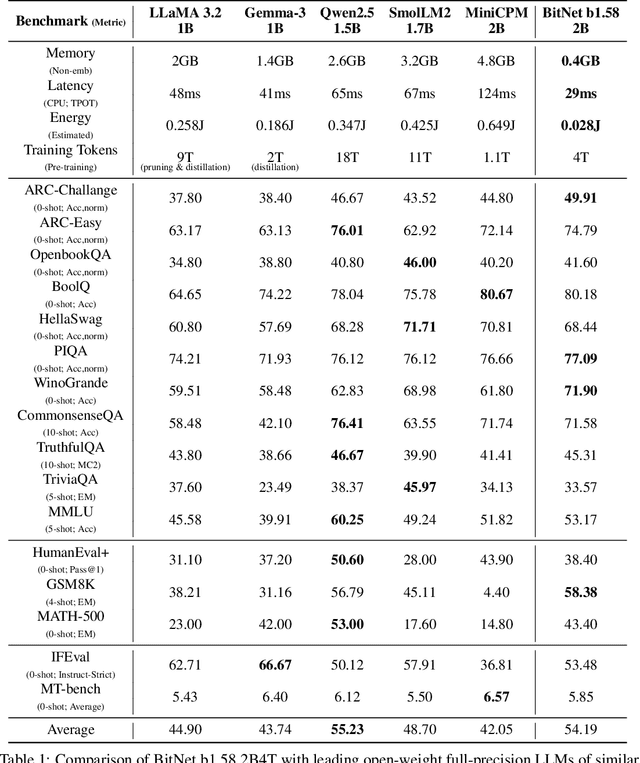
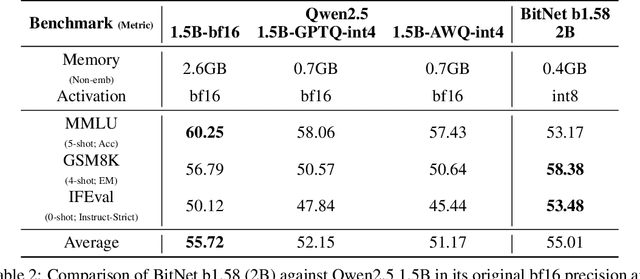
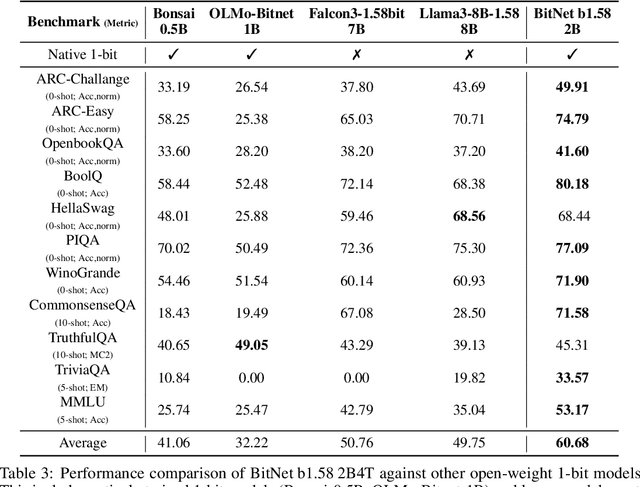
Abstract:We introduce BitNet b1.58 2B4T, the first open-source, native 1-bit Large Language Model (LLM) at the 2-billion parameter scale. Trained on a corpus of 4 trillion tokens, the model has been rigorously evaluated across benchmarks covering language understanding, mathematical reasoning, coding proficiency, and conversational ability. Our results demonstrate that BitNet b1.58 2B4T achieves performance on par with leading open-weight, full-precision LLMs of similar size, while offering significant advantages in computational efficiency, including substantially reduced memory footprint, energy consumption, and decoding latency. To facilitate further research and adoption, the model weights are released via Hugging Face along with open-source inference implementations for both GPU and CPU architectures.
GDiffRetro: Retrosynthesis Prediction with Dual Graph Enhanced Molecular Representation and Diffusion Generation
Jan 14, 2025



Abstract:Retrosynthesis prediction focuses on identifying reactants capable of synthesizing a target product. Typically, the retrosynthesis prediction involves two phases: Reaction Center Identification and Reactant Generation. However, we argue that most existing methods suffer from two limitations in the two phases: (i) Existing models do not adequately capture the ``face'' information in molecular graphs for the reaction center identification. (ii) Current approaches for the reactant generation predominantly use sequence generation in a 2D space, which lacks versatility in generating reasonable distributions for completed reactive groups and overlooks molecules' inherent 3D properties. To overcome the above limitations, we propose GDiffRetro. For the reaction center identification, GDiffRetro uniquely integrates the original graph with its corresponding dual graph to represent molecular structures, which helps guide the model to focus more on the faces in the graph. For the reactant generation, GDiffRetro employs a conditional diffusion model in 3D to further transform the obtained synthon into a complete reactant. Our experimental findings reveal that GDiffRetro outperforms state-of-the-art semi-template models across various evaluative metrics.
Med-2E3: A 2D-Enhanced 3D Medical Multimodal Large Language Model
Nov 19, 2024



Abstract:The analysis of 3D medical images is crucial for modern healthcare, yet traditional task-specific models are becoming increasingly inadequate due to limited generalizability across diverse clinical scenarios. Multimodal large language models (MLLMs) offer a promising solution to these challenges. However, existing MLLMs have limitations in fully leveraging the rich, hierarchical information embedded in 3D medical images. Inspired by clinical practice, where radiologists focus on both 3D spatial structure and 2D planar content, we propose Med-2E3, a novel MLLM for 3D medical image analysis that integrates 3D and 2D encoders. To aggregate 2D features more effectively, we design a Text-Guided Inter-Slice (TG-IS) scoring module, which scores the attention of each 2D slice based on slice contents and task instructions. To the best of our knowledge, Med-2E3 is the first MLLM to integrate both 3D and 2D features for 3D medical image analysis. Experiments on a large-scale, open-source 3D medical multimodal benchmark demonstrate that Med-2E3 exhibits task-specific attention distribution and significantly outperforms current state-of-the-art models, with a 14% improvement in report generation and a 5% gain in medical visual question answering (VQA), highlighting the model's potential in addressing complex multimodal clinical tasks. The code will be released upon acceptance.
DiffuseST: Unleashing the Capability of the Diffusion Model for Style Transfer
Oct 19, 2024
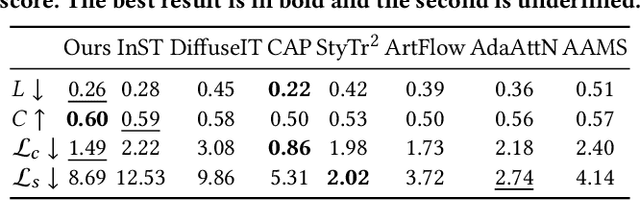
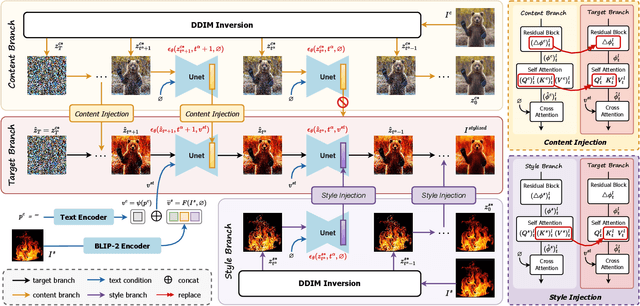
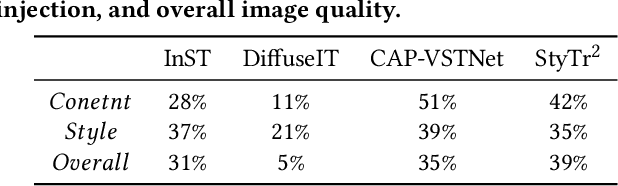
Abstract:Style transfer aims to fuse the artistic representation of a style image with the structural information of a content image. Existing methods train specific networks or utilize pre-trained models to learn content and style features. However, they rely solely on textual or spatial representations that are inadequate to achieve the balance between content and style. In this work, we propose a novel and training-free approach for style transfer, combining textual embedding with spatial features and separating the injection of content or style. Specifically, we adopt the BLIP-2 encoder to extract the textual representation of the style image. We utilize the DDIM inversion technique to extract intermediate embeddings in content and style branches as spatial features. Finally, we harness the step-by-step property of diffusion models by separating the injection of content and style in the target branch, which improves the balance between content preservation and style fusion. Various experiments have demonstrated the effectiveness and robustness of our proposed DiffeseST for achieving balanced and controllable style transfer results, as well as the potential to extend to other tasks.
Magnet: We Never Know How Text-to-Image Diffusion Models Work, Until We Learn How Vision-Language Models Function
Sep 30, 2024



Abstract:Text-to-image diffusion models particularly Stable Diffusion, have revolutionized the field of computer vision. However, the synthesis quality often deteriorates when asked to generate images that faithfully represent complex prompts involving multiple attributes and objects. While previous studies suggest that blended text embeddings lead to improper attribute binding, few have explored this in depth. In this work, we critically examine the limitations of the CLIP text encoder in understanding attributes and investigate how this affects diffusion models. We discern a phenomenon of attribute bias in the text space and highlight a contextual issue in padding embeddings that entangle different concepts. We propose \textbf{Magnet}, a novel training-free approach to tackle the attribute binding problem. We introduce positive and negative binding vectors to enhance disentanglement, further with a neighbor strategy to increase accuracy. Extensive experiments show that Magnet significantly improves synthesis quality and binding accuracy with negligible computational cost, enabling the generation of unconventional and unnatural concepts.
Uni-Med: A Unified Medical Generalist Foundation Model For Multi-Task Learning Via Connector-MoE
Sep 26, 2024



Abstract:Multi-modal large language models (MLLMs) have shown impressive capabilities as a general-purpose interface for various visual and linguistic tasks. However, building a unified MLLM for multi-task learning in the medical field remains a thorny challenge. To mitigate the tug-of-war problem of multi-modal multi-task optimization, recent advances primarily focus on improving the LLM components, while neglecting the connector that bridges the gap between modalities. In this paper, we introduce Uni-Med, a novel medical generalist foundation model which consists of a universal visual feature extraction module, a connector mixture-of-experts (CMoE) module, and an LLM. Benefiting from the proposed CMoE that leverages a well-designed router with a mixture of projection experts at the connector, Uni-Med achieves efficient solution to the tug-of-war problem and can perform six different medical tasks including question answering, visual question answering, report generation, referring expression comprehension, referring expression generation and image classification. To the best of our knowledge, Uni-Med is the first effort to tackle multi-task interference at the connector. Extensive ablation experiments validate the effectiveness of introducing CMoE under any configuration, with up to an average 8% performance gains. We further provide interpretation analysis of the tug-of-war problem from the perspective of gradient optimization and parameter statistics. Compared to previous state-of-the-art medical MLLMs, Uni-Med achieves competitive or superior evaluation metrics on diverse tasks. Code, data and model will be soon available at GitHub.
SparseSSP: 3D Subcellular Structure Prediction from Sparse-View Transmitted Light Images
Jul 03, 2024



Abstract:Traditional fluorescence staining is phototoxic to live cells, slow, and expensive; thus, the subcellular structure prediction (SSP) from transmitted light (TL) images is emerging as a label-free, faster, low-cost alternative. However, existing approaches utilize 3D networks for one-to-one voxel level dense prediction, which necessitates a frequent and time-consuming Z-axis imaging process. Moreover, 3D convolutions inevitably lead to significant computation and GPU memory overhead. Therefore, we propose an efficient framework, SparseSSP, predicting fluorescent intensities within the target voxel grid in an efficient paradigm instead of relying entirely on 3D topologies. In particular, SparseSSP makes two pivotal improvements to prior works. First, SparseSSP introduces a one-to-many voxel mapping paradigm, which permits the sparse TL slices to reconstruct the subcellular structure. Secondly, we propose a hybrid dimensions topology, which folds the Z-axis information into channel features, enabling the 2D network layers to tackle SSP under low computational cost. We conduct extensive experiments to validate the effectiveness and advantages of SparseSSP on diverse sparse imaging ratios, and our approach achieves a leading performance compared to pure 3D topologies. SparseSSP reduces imaging frequencies compared to previous dense-view SSP (i.e., the number of imaging is reduced up to 87.5% at most), which is significant in visualizing rapid biological dynamics on low-cost devices and samples.
Ultrasound Report Generation with Cross-Modality Feature Alignment via Unsupervised Guidance
Jun 02, 2024Abstract:Automatic report generation has arisen as a significant research area in computer-aided diagnosis, aiming to alleviate the burden on clinicians by generating reports automatically based on medical images. In this work, we propose a novel framework for automatic ultrasound report generation, leveraging a combination of unsupervised and supervised learning methods to aid the report generation process. Our framework incorporates unsupervised learning methods to extract potential knowledge from ultrasound text reports, serving as the prior information to guide the model in aligning visual and textual features, thereby addressing the challenge of feature discrepancy. Additionally, we design a global semantic comparison mechanism to enhance the performance of generating more comprehensive and accurate medical reports. To enable the implementation of ultrasound report generation, we constructed three large-scale ultrasound image-text datasets from different organs for training and validation purposes. Extensive evaluations with other state-of-the-art approaches exhibit its superior performance across all three datasets. Code and dataset are valuable at this link.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge