Xuecang Zhang
GDiffRetro: Retrosynthesis Prediction with Dual Graph Enhanced Molecular Representation and Diffusion Generation
Jan 14, 2025



Abstract:Retrosynthesis prediction focuses on identifying reactants capable of synthesizing a target product. Typically, the retrosynthesis prediction involves two phases: Reaction Center Identification and Reactant Generation. However, we argue that most existing methods suffer from two limitations in the two phases: (i) Existing models do not adequately capture the ``face'' information in molecular graphs for the reaction center identification. (ii) Current approaches for the reactant generation predominantly use sequence generation in a 2D space, which lacks versatility in generating reasonable distributions for completed reactive groups and overlooks molecules' inherent 3D properties. To overcome the above limitations, we propose GDiffRetro. For the reaction center identification, GDiffRetro uniquely integrates the original graph with its corresponding dual graph to represent molecular structures, which helps guide the model to focus more on the faces in the graph. For the reactant generation, GDiffRetro employs a conditional diffusion model in 3D to further transform the obtained synthon into a complete reactant. Our experimental findings reveal that GDiffRetro outperforms state-of-the-art semi-template models across various evaluative metrics.
FusionANNS: An Efficient CPU/GPU Cooperative Processing Architecture for Billion-scale Approximate Nearest Neighbor Search
Sep 25, 2024



Abstract:Approximate nearest neighbor search (ANNS) has emerged as a crucial component of database and AI infrastructure. Ever-increasing vector datasets pose significant challenges in terms of performance, cost, and accuracy for ANNS services. None of modern ANNS systems can address these issues simultaneously. We present FusionANNS, a high-throughput, low-latency, cost-efficient, and high-accuracy ANNS system for billion-scale datasets using SSDs and only one entry-level GPU. The key idea of FusionANNS lies in CPU/GPU collaborative filtering and re-ranking mechanisms, which significantly reduce I/O operations across CPUs, GPU, and SSDs to break through the I/O performance bottleneck. Specifically, we propose three novel designs: (1) multi-tiered indexing to avoid data swapping between CPUs and GPU, (2) heuristic re-ranking to eliminate unnecessary I/Os and computations while guaranteeing high accuracy, and (3) redundant-aware I/O deduplication to further improve I/O efficiency. We implement FusionANNS and compare it with the state-of-the-art SSD-based ANNS system--SPANN and GPU-accelerated in-memory ANNS system--RUMMY. Experimental results show that FusionANNS achieves 1) 9.4-13.1X higher query per second (QPS) and 5.7-8.8X higher cost efficiency compared with SPANN; 2) and 2-4.9X higher QPS and 2.3-6.8X higher cost efficiency compared with RUMMY, while guaranteeing low latency and high accuracy.
Sculpting Molecules in 3D: A Flexible Substructure Aware Framework for Text-Oriented Molecular Optimization
Mar 06, 2024



Abstract:The integration of deep learning, particularly AI-Generated Content, with high-quality data derived from ab initio calculations has emerged as a promising avenue for transforming the landscape of scientific research. However, the challenge of designing molecular drugs or materials that incorporate multi-modality prior knowledge remains a critical and complex undertaking. Specifically, achieving a practical molecular design necessitates not only meeting the diversity requirements but also addressing structural and textural constraints with various symmetries outlined by domain experts. In this article, we present an innovative approach to tackle this inverse design problem by formulating it as a multi-modality guidance generation/optimization task. Our proposed solution involves a textural-structure alignment symmetric diffusion framework for the implementation of molecular generation/optimization tasks, namely 3DToMolo. 3DToMolo aims to harmonize diverse modalities, aligning them seamlessly to produce molecular structures adhere to specified symmetric structural and textural constraints by experts in the field. Experimental trials across three guidance generation settings have shown a superior hit generation performance compared to state-of-the-art methodologies. Moreover, 3DToMolo demonstrates the capability to generate novel molecules, incorporating specified target substructures, without the need for prior knowledge. This work not only holds general significance for the advancement of deep learning methodologies but also paves the way for a transformative shift in molecular design strategies. 3DToMolo creates opportunities for a more nuanced and effective exploration of the vast chemical space, opening new frontiers in the development of molecular entities with tailored properties and functionalities.
Molecule Joint Auto-Encoding: Trajectory Pretraining with 2D and 3D Diffusion
Dec 06, 2023Abstract:Recently, artificial intelligence for drug discovery has raised increasing interest in both machine learning and chemistry domains. The fundamental building block for drug discovery is molecule geometry and thus, the molecule's geometrical representation is the main bottleneck to better utilize machine learning techniques for drug discovery. In this work, we propose a pretraining method for molecule joint auto-encoding (MoleculeJAE). MoleculeJAE can learn both the 2D bond (topology) and 3D conformation (geometry) information, and a diffusion process model is applied to mimic the augmented trajectories of such two modalities, based on which, MoleculeJAE will learn the inherent chemical structure in a self-supervised manner. Thus, the pretrained geometrical representation in MoleculeJAE is expected to benefit downstream geometry-related tasks. Empirically, MoleculeJAE proves its effectiveness by reaching state-of-the-art performance on 15 out of 20 tasks by comparing it with 12 competitive baselines.
NeutronStream: A Dynamic GNN Training Framework with Sliding Window for Graph Streams
Dec 05, 2023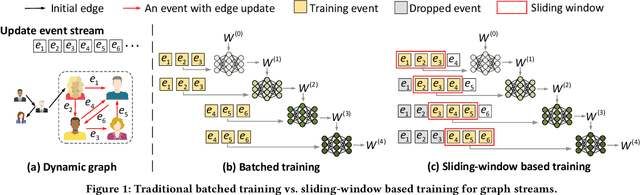
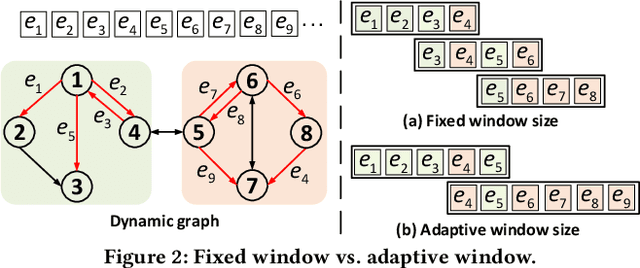
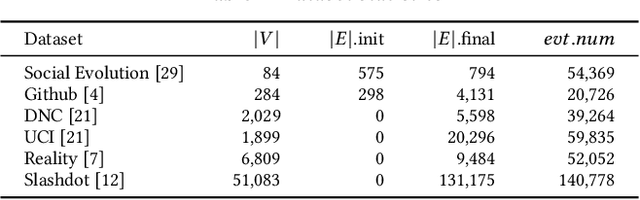
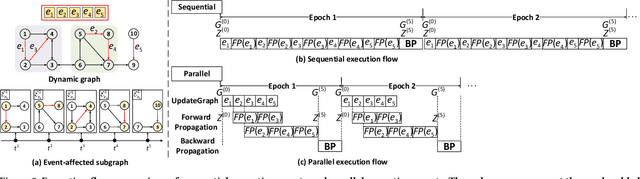
Abstract:Existing Graph Neural Network (GNN) training frameworks have been designed to help developers easily create performant GNN implementations. However, most existing GNN frameworks assume that the input graphs are static, but ignore that most real-world graphs are constantly evolving. Though many dynamic GNN models have emerged to learn from evolving graphs, the training process of these dynamic GNNs is dramatically different from traditional GNNs in that it captures both the spatial and temporal dependencies of graph updates. This poses new challenges for designing dynamic GNN training frameworks. First, the traditional batched training method fails to capture real-time structural evolution information. Second, the time-dependent nature makes parallel training hard to design. Third, it lacks system supports for users to efficiently implement dynamic GNNs. In this paper, we present NeutronStream, a framework for training dynamic GNN models. NeutronStream abstracts the input dynamic graph into a chronologically updated stream of events and processes the stream with an optimized sliding window to incrementally capture the spatial-temporal dependencies of events. Furthermore, NeutronStream provides a parallel execution engine to tackle the sequential event processing challenge to achieve high performance. NeutronStream also integrates a built-in graph storage structure that supports dynamic updates and provides a set of easy-to-use APIs that allow users to express their dynamic GNNs. Our experimental results demonstrate that, compared to state-of-the-art dynamic GNN implementations, NeutronStream achieves speedups ranging from 1.48X to 5.87X and an average accuracy improvement of 3.97%.
Large Language Models as Topological Structure Enhancers for Text-Attributed Graphs
Nov 24, 2023



Abstract:The latest advancements in large language models (LLMs) have revolutionized the field of natural language processing (NLP). Inspired by the success of LLMs in NLP tasks, some recent work has begun investigating the potential of applying LLMs in graph learning tasks. However, most of the existing work focuses on utilizing LLMs as powerful node feature augmenters, leaving employing LLMs to enhance graph topological structures an understudied problem. In this work, we explore how to leverage the information retrieval and text generation capabilities of LLMs to refine/enhance the topological structure of text-attributed graphs (TAGs) under the node classification setting. First, we propose using LLMs to help remove unreliable edges and add reliable ones in the TAG. Specifically, we first let the LLM output the semantic similarity between node attributes through delicate prompt designs, and then perform edge deletion and edge addition based on the similarity. Second, we propose using pseudo-labels generated by the LLM to improve graph topology, that is, we introduce the pseudo-label propagation as a regularization to guide the graph neural network (GNN) in learning proper edge weights. Finally, we incorporate the two aforementioned LLM-based methods for graph topological refinement into the process of GNN training, and perform extensive experiments on four real-world datasets. The experimental results demonstrate the effectiveness of LLM-based graph topology refinement (achieving a 0.15%--2.47% performance gain on public benchmarks).
Empower Text-Attributed Graphs Learning with Large Language Models (LLMs)
Oct 15, 2023



Abstract:Text-attributed graphs have recently garnered significant attention due to their wide range of applications in web domains. Existing methodologies employ word embedding models for acquiring text representations as node features, which are subsequently fed into Graph Neural Networks (GNNs) for training. Recently, the advent of Large Language Models (LLMs) has introduced their powerful capabilities in information retrieval and text generation, which can greatly enhance the text attributes of graph data. Furthermore, the acquisition and labeling of extensive datasets are both costly and time-consuming endeavors. Consequently, few-shot learning has emerged as a crucial problem in the context of graph learning tasks. In order to tackle this challenge, we propose a lightweight paradigm called ENG, which adopts a plug-and-play approach to empower text-attributed graphs through node generation using LLMs. Specifically, we utilize LLMs to extract semantic information from the labels and generate samples that belong to these categories as exemplars. Subsequently, we employ an edge predictor to capture the structural information inherent in the raw dataset and integrate the newly generated samples into the original graph. This approach harnesses LLMs for enhancing class-level information and seamlessly introduces labeled nodes and edges without modifying the raw dataset, thereby facilitating the node classification task in few-shot scenarios. Extensive experiments demonstrate the outstanding performance of our proposed paradigm, particularly in low-shot scenarios. For instance, in the 1-shot setting of the ogbn-arxiv dataset, ENG achieves a 76% improvement over the baseline model.
DiskANN++: Efficient Page-based Search over Isomorphic Mapped Graph Index using Query-sensitivity Entry Vertex
Sep 30, 2023Abstract:Given a vector dataset $\mathcal{X}$ and a query vector $\vec{x}_q$, graph-based Approximate Nearest Neighbor Search (ANNS) aims to build a graph index $G$ and approximately return vectors with minimum distances to $\vec{x}_q$ by searching over $G$. The main drawback of graph-based ANNS is that a graph index would be too large to fit into the memory especially for a large-scale $\mathcal{X}$. To solve this, a Product Quantization (PQ)-based hybrid method called DiskANN is proposed to store a low-dimensional PQ index in memory and retain a graph index in SSD, thus reducing memory overhead while ensuring a high search accuracy. However, it suffers from two I/O issues that significantly affect the overall efficiency: (1) long routing path from an entry vertex to the query's neighborhood that results in large number of I/O requests and (2) redundant I/O requests during the routing process. We propose an optimized DiskANN++ to overcome above issues. Specifically, for the first issue, we present a query-sensitive entry vertex selection strategy to replace DiskANN's static graph-central entry vertex by a dynamically determined entry vertex that is close to the query. For the second I/O issue, we present an isomorphic mapping on DiskANN's graph index to optimize the SSD layout and propose an asynchronously optimized Pagesearch based on the optimized SSD layout as an alternative to DiskANN's beamsearch. Comprehensive experimental studies on eight real-world datasets demonstrate our DiskANN++'s superiority on efficiency. We achieve a notable 1.5 X to 2.2 X improvement on QPS compared to DiskANN, given the same accuracy constraint.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge