Jintu Zheng
FTP: A Fine-grained Token-wise Pruner for Large Language Models via Token Routing
Dec 16, 2024Abstract:Recently, large language models (LLMs) have demonstrated superior performance across various tasks by adhering to scaling laws, which significantly increase model size. However, the huge computation overhead during inference hinders the deployment in industrial applications. Many works leverage traditional compression approaches to boost model inference, but these always introduce additional training costs to restore the performance and the pruning results typically show noticeable performance drops compared to the original model when aiming for a specific level of acceleration. To address these issues, we propose a fine-grained token-wise pruning approach for the LLMs, which presents a learnable router to adaptively identify the less important tokens and skip them across model blocks to reduce computational cost during inference. To construct the router efficiently, we present a search-based sparsity scheduler for pruning sparsity allocation, a trainable router combined with our proposed four low-dimensional factors as input and three proposed losses. We conduct extensive experiments across different benchmarks on different LLMs to demonstrate the superiority of our method. Our approach achieves state-of-the-art (SOTA) pruning results, surpassing other existing pruning methods. For instance, our method outperforms BlockPruner and ShortGPT by approximately 10 points on both LLaMA2-7B and Qwen1.5-7B in accuracy retention at comparable token sparsity levels.
DiP-GO: A Diffusion Pruner via Few-step Gradient Optimization
Oct 22, 2024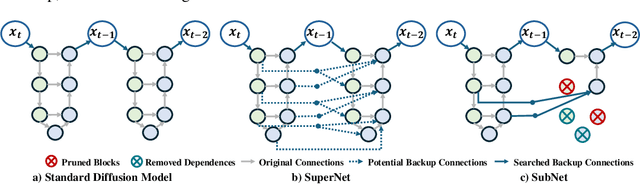
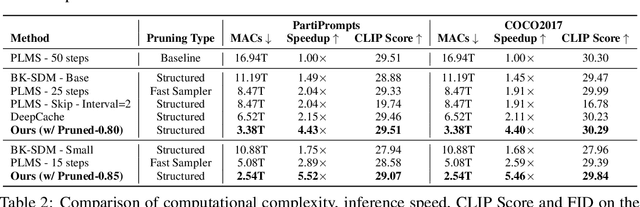
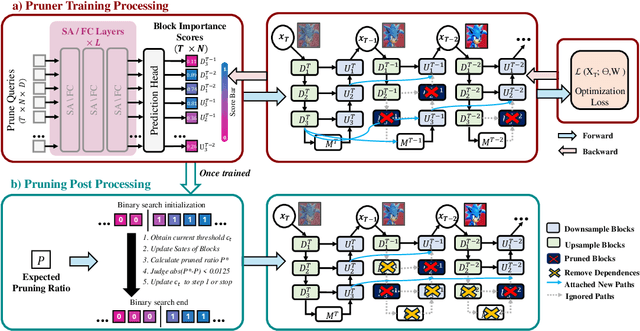
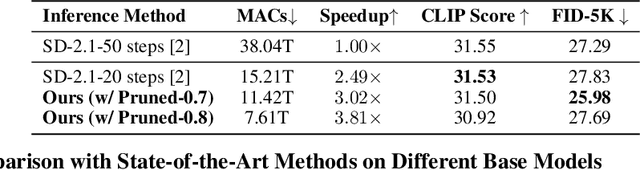
Abstract:Diffusion models have achieved remarkable progress in the field of image generation due to their outstanding capabilities. However, these models require substantial computing resources because of the multi-step denoising process during inference. While traditional pruning methods have been employed to optimize these models, the retraining process necessitates large-scale training datasets and extensive computational costs to maintain generalization ability, making it neither convenient nor efficient. Recent studies attempt to utilize the similarity of features across adjacent denoising stages to reduce computational costs through simple and static strategies. However, these strategies cannot fully harness the potential of the similar feature patterns across adjacent timesteps. In this work, we propose a novel pruning method that derives an efficient diffusion model via a more intelligent and differentiable pruner. At the core of our approach is casting the model pruning process into a SubNet search process. Specifically, we first introduce a SuperNet based on standard diffusion via adding some backup connections built upon the similar features. We then construct a plugin pruner network and design optimization losses to identify redundant computation. Finally, our method can identify an optimal SubNet through few-step gradient optimization and a simple post-processing procedure. We conduct extensive experiments on various diffusion models including Stable Diffusion series and DiTs. Our DiP-GO approach achieves 4.4 x speedup for SD-1.5 without any loss of accuracy, significantly outperforming the previous state-of-the-art methods.
Memory Matching is not Enough: Jointly Improving Memory Matching and Decoding for Video Object Segmentation
Sep 22, 2024Abstract:Memory-based video object segmentation methods model multiple objects over long temporal-spatial spans by establishing memory bank, which achieve the remarkable performance. However, they struggle to overcome the false matching and are prone to lose critical information, resulting in confusion among different objects. In this paper, we propose an effective approach which jointly improving the matching and decoding stages to alleviate the false matching issue.For the memory matching stage, we present a cost aware mechanism that suppresses the slight errors for short-term memory and a shunted cross-scale matching for long-term memory which establish a wide filed matching spaces for various object scales. For the readout decoding stage, we implement a compensatory mechanism aims at recovering the essential information where missing at the matching stage. Our approach achieves the outstanding performance in several popular benchmarks (i.e., DAVIS 2016&2017 Val (92.4%&88.1%), and DAVIS 2017 Test (83.9%)), and achieves 84.8%&84.6% on YouTubeVOS 2018&2019 Val.
SparseSSP: 3D Subcellular Structure Prediction from Sparse-View Transmitted Light Images
Jul 03, 2024



Abstract:Traditional fluorescence staining is phototoxic to live cells, slow, and expensive; thus, the subcellular structure prediction (SSP) from transmitted light (TL) images is emerging as a label-free, faster, low-cost alternative. However, existing approaches utilize 3D networks for one-to-one voxel level dense prediction, which necessitates a frequent and time-consuming Z-axis imaging process. Moreover, 3D convolutions inevitably lead to significant computation and GPU memory overhead. Therefore, we propose an efficient framework, SparseSSP, predicting fluorescent intensities within the target voxel grid in an efficient paradigm instead of relying entirely on 3D topologies. In particular, SparseSSP makes two pivotal improvements to prior works. First, SparseSSP introduces a one-to-many voxel mapping paradigm, which permits the sparse TL slices to reconstruct the subcellular structure. Secondly, we propose a hybrid dimensions topology, which folds the Z-axis information into channel features, enabling the 2D network layers to tackle SSP under low computational cost. We conduct extensive experiments to validate the effectiveness and advantages of SparseSSP on diverse sparse imaging ratios, and our approach achieves a leading performance compared to pure 3D topologies. SparseSSP reduces imaging frequencies compared to previous dense-view SSP (i.e., the number of imaging is reduced up to 87.5% at most), which is significant in visualizing rapid biological dynamics on low-cost devices and samples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge