Zhaohu Xing
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
Toward Real-World High-Precision Image Matting and Segmentation
Jan 17, 2026Abstract:High-precision scene parsing tasks, including image matting and dichotomous segmentation, aim to accurately predict masks with extremely fine details (such as hair). Most existing methods focus on salient, single foreground objects. While interactive methods allow for target adjustment, their class-agnostic design restricts generalization across different categories. Furthermore, the scarcity of high-quality annotation has led to a reliance on inharmonious synthetic data, resulting in poor generalization to real-world scenarios. To this end, we propose a Foreground Consistent Learning model, dubbed as FCLM, to address the aforementioned issues. Specifically, we first introduce a Depth-Aware Distillation strategy where we transfer the depth-related knowledge for better foreground representation. Considering the data dilemma, we term the processing of synthetic data as domain adaptation problem where we propose a domain-invariant learning strategy to focus on foreground learning. To support interactive prediction, we contribute an Object-Oriented Decoder that can receive both visual and language prompts to predict the referring target. Experimental results show that our method quantitatively and qualitatively outperforms SOTA methods.
VQ-Seg: Vector-Quantized Token Perturbation for Semi-Supervised Medical Image Segmentation
Jan 15, 2026Abstract:Consistency learning with feature perturbation is a widely used strategy in semi-supervised medical image segmentation. However, many existing perturbation methods rely on dropout, and thus require a careful manual tuning of the dropout rate, which is a sensitive hyperparameter and often difficult to optimize and may lead to suboptimal regularization. To overcome this limitation, we propose VQ-Seg, the first approach to employ vector quantization (VQ) to discretize the feature space and introduce a novel and controllable Quantized Perturbation Module (QPM) that replaces dropout. Our QPM perturbs discrete representations by shuffling the spatial locations of codebook indices, enabling effective and controllable regularization. To mitigate potential information loss caused by quantization, we design a dual-branch architecture where the post-quantization feature space is shared by both image reconstruction and segmentation tasks. Moreover, we introduce a Post-VQ Feature Adapter (PFA) to incorporate guidance from a foundation model (FM), supplementing the high-level semantic information lost during quantization. Furthermore, we collect a large-scale Lung Cancer (LC) dataset comprising 828 CT scans annotated for central-type lung carcinoma. Extensive experiments on the LC dataset and other public benchmarks demonstrate the effectiveness of our method, which outperforms state-of-the-art approaches. Code available at: https://github.com/script-Yang/VQ-Seg.
K-Stain: Keypoint-Driven Correspondence for H&E-to-IHC Virtual Staining
Nov 10, 2025Abstract:Virtual staining offers a promising method for converting Hematoxylin and Eosin (H&E) images into Immunohistochemical (IHC) images, eliminating the need for costly chemical processes. However, existing methods often struggle to utilize spatial information effectively due to misalignment in tissue slices. To overcome this challenge, we leverage keypoints as robust indicators of spatial correspondence, enabling more precise alignment and integration of structural details in synthesized IHC images. We introduce K-Stain, a novel framework that employs keypoint-based spatial and semantic relationships to enhance synthesized IHC image fidelity. K-Stain comprises three main components: (1) a Hierarchical Spatial Keypoint Detector (HSKD) for identifying keypoints in stain images, (2) a Keypoint-aware Enhancement Generator (KEG) that integrates these keypoints during image generation, and (3) a Keypoint Guided Discriminator (KGD) that improves the discriminator's sensitivity to spatial details. Our approach leverages contextual information from adjacent slices, resulting in more accurate and visually consistent IHC images. Extensive experiments show that K-Stain outperforms state-of-the-art methods in quantitative metrics and visual quality.
Toward Medical Deepfake Detection: A Comprehensive Dataset and Novel Method
Sep 19, 2025Abstract:The rapid advancement of generative AI in medical imaging has introduced both significant opportunities and serious challenges, especially the risk that fake medical images could undermine healthcare systems. These synthetic images pose serious risks, such as diagnostic deception, financial fraud, and misinformation. However, research on medical forensics to counter these threats remains limited, and there is a critical lack of comprehensive datasets specifically tailored for this field. Additionally, existing media forensic methods, which are primarily designed for natural or facial images, are inadequate for capturing the distinct characteristics and subtle artifacts of AI-generated medical images. To tackle these challenges, we introduce \textbf{MedForensics}, a large-scale medical forensics dataset encompassing six medical modalities and twelve state-of-the-art medical generative models. We also propose \textbf{DSKI}, a novel \textbf{D}ual-\textbf{S}tage \textbf{K}nowledge \textbf{I}nfusing detector that constructs a vision-language feature space tailored for the detection of AI-generated medical images. DSKI comprises two core components: 1) a cross-domain fine-trace adapter (CDFA) for extracting subtle forgery clues from both spatial and noise domains during training, and 2) a medical forensic retrieval module (MFRM) that boosts detection accuracy through few-shot retrieval during testing. Experimental results demonstrate that DSKI significantly outperforms both existing methods and human experts, achieving superior accuracy across multiple medical modalities.
HybridMamba: A Dual-domain Mamba for 3D Medical Image Segmentation
Sep 18, 2025Abstract:In the domain of 3D biomedical image segmentation, Mamba exhibits the superior performance for it addresses the limitations in modeling long-range dependencies inherent to CNNs and mitigates the abundant computational overhead associated with Transformer-based frameworks when processing high-resolution medical volumes. However, attaching undue importance to global context modeling may inadvertently compromise critical local structural information, thus leading to boundary ambiguity and regional distortion in segmentation outputs. Therefore, we propose the HybridMamba, an architecture employing dual complementary mechanisms: 1) a feature scanning strategy that progressively integrates representations both axial-traversal and local-adaptive pathways to harmonize the relationship between local and global representations, and 2) a gated module combining spatial-frequency analysis for comprehensive contextual modeling. Besides, we collect a multi-center CT dataset related to lung cancer. Experiments on MRI and CT datasets demonstrate that HybridMamba significantly outperforms the state-of-the-art methods in 3D medical image segmentation.
Technical Report of TeleChat2, TeleChat2.5 and T1
Jul 24, 2025
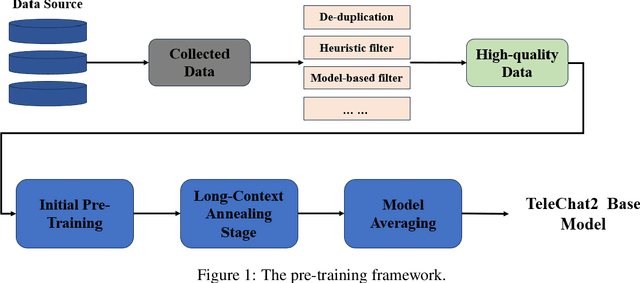
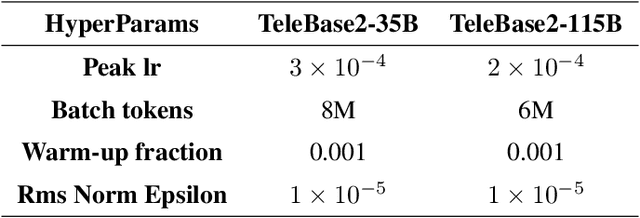
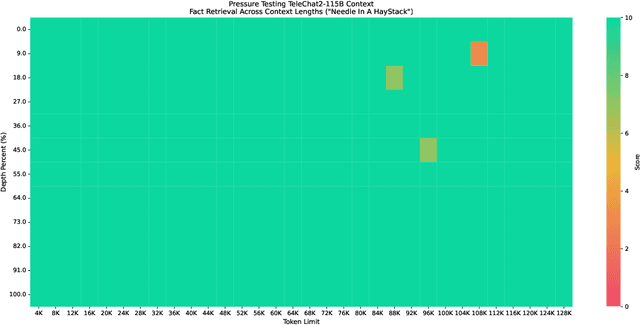
Abstract:We introduce the latest series of TeleChat models: \textbf{TeleChat2}, \textbf{TeleChat2.5}, and \textbf{T1}, offering a significant upgrade over their predecessor, TeleChat. Despite minimal changes to the model architecture, the new series achieves substantial performance gains through enhanced training strategies in both pre-training and post-training stages. The series begins with \textbf{TeleChat2}, which undergoes pretraining on 10 trillion high-quality and diverse tokens. This is followed by Supervised Fine-Tuning (SFT) and Direct Preference Optimization (DPO) to further enhance its capabilities. \textbf{TeleChat2.5} and \textbf{T1} expand the pipeline by incorporating a continual pretraining phase with domain-specific datasets, combined with reinforcement learning (RL) to improve performance in code generation and mathematical reasoning tasks. The \textbf{T1} variant is designed for complex reasoning, supporting long Chain-of-Thought (CoT) reasoning and demonstrating substantial improvements in mathematics and coding. In contrast, \textbf{TeleChat2.5} prioritizes speed, delivering rapid inference. Both flagship models of \textbf{T1} and \textbf{TeleChat2.5} are dense Transformer-based architectures with 115B parameters, showcasing significant advancements in reasoning and general task performance compared to the original TeleChat. Notably, \textbf{T1-115B} outperform proprietary models such as OpenAI's o1-mini and GPT-4o. We publicly release \textbf{TeleChat2}, \textbf{TeleChat2.5} and \textbf{T1}, including post-trained versions with 35B and 115B parameters, to empower developers and researchers with state-of-the-art language models tailored for diverse applications.
PosterCraft: Rethinking High-Quality Aesthetic Poster Generation in a Unified Framework
Jun 12, 2025
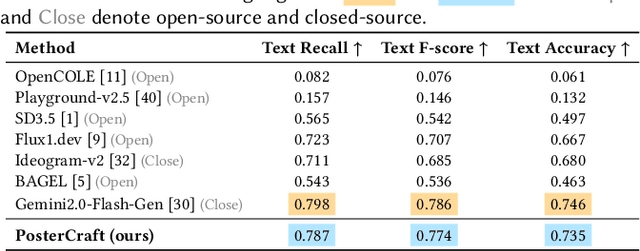
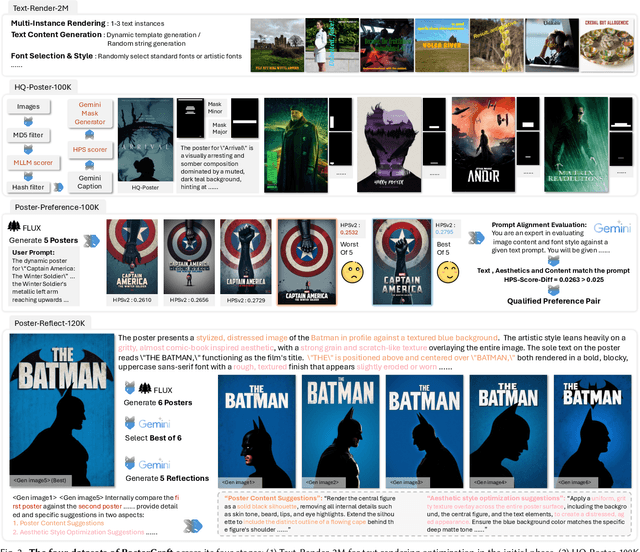
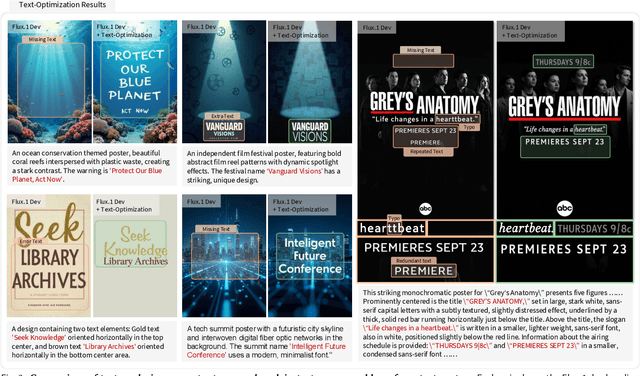
Abstract:Generating aesthetic posters is more challenging than simple design images: it requires not only precise text rendering but also the seamless integration of abstract artistic content, striking layouts, and overall stylistic harmony. To address this, we propose PosterCraft, a unified framework that abandons prior modular pipelines and rigid, predefined layouts, allowing the model to freely explore coherent, visually compelling compositions. PosterCraft employs a carefully designed, cascaded workflow to optimize the generation of high-aesthetic posters: (i) large-scale text-rendering optimization on our newly introduced Text-Render-2M dataset; (ii) region-aware supervised fine-tuning on HQ-Poster100K; (iii) aesthetic-text-reinforcement learning via best-of-n preference optimization; and (iv) joint vision-language feedback refinement. Each stage is supported by a fully automated data-construction pipeline tailored to its specific needs, enabling robust training without complex architectural modifications. Evaluated on multiple experiments, PosterCraft significantly outperforms open-source baselines in rendering accuracy, layout coherence, and overall visual appeal-approaching the quality of SOTA commercial systems. Our code, models, and datasets can be found in the Project page: https://ephemeral182.github.io/PosterCraft
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
Infinity-MM: Scaling Multimodal Performance with Large-Scale and High-Quality Instruction Data
Oct 24, 2024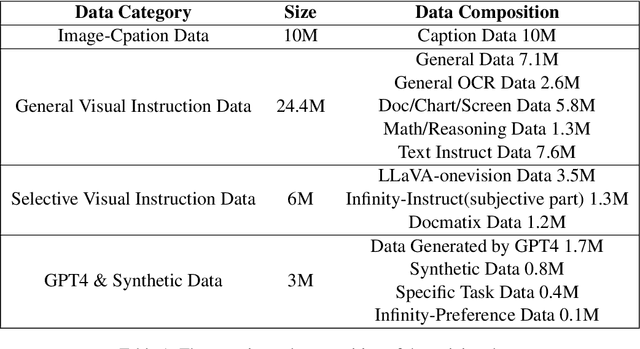
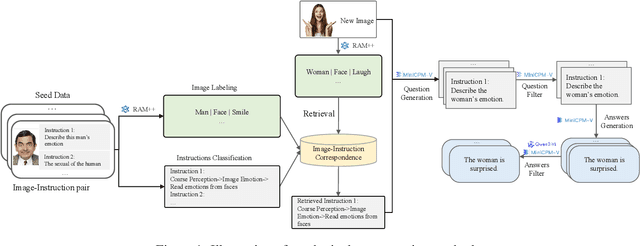
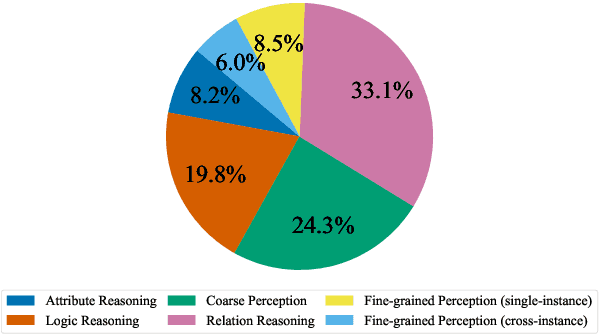
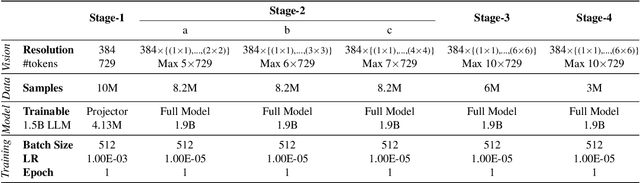
Abstract:Vision-Language Models (VLMs) have recently made significant progress, but the limited scale and quality of open-source instruction data hinder their performance compared to closed-source models. In this work, we address this limitation by introducing Infinity-MM, a large-scale multimodal instruction dataset with 40 million samples, enhanced through rigorous quality filtering and deduplication. We also propose a synthetic instruction generation method based on open-source VLMs, using detailed image annotations and diverse question generation. Using this data, we trained a 2-billion-parameter VLM, Aquila-VL-2B, achieving state-of-the-art (SOTA) performance for models of similar scale. This demonstrates that expanding instruction data and generating synthetic data can significantly improve the performance of open-source models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge