Xiaoyang Huang
InstantSticker: Realistic Decal Blending via Disentangled Object Reconstruction
Apr 09, 2025



Abstract:We present InstantSticker, a disentangled reconstruction pipeline based on Image-Based Lighting (IBL), which focuses on highly realistic decal blending, simulates stickers attached to the reconstructed surface, and allows for instant editing and real-time rendering. To achieve stereoscopic impression of the decal, we introduce shadow factor into IBL, which can be adaptively optimized during training. This allows the shadow brightness of surfaces to be accurately decomposed rather than baked into the diffuse color, ensuring that the edited texture exhibits authentic shading. To address the issues of warping and blurriness in previous methods, we apply As-Rigid-As-Possible (ARAP) parameterization to pre-unfold a specified area of the mesh and use the local UV mapping combined with a neural texture map to enhance the ability to express high-frequency details in that area. For instant editing, we utilize the Disney BRDF model, explicitly defining material colors with 3-channel diffuse albedo. This enables instant replacement of albedo RGB values during the editing process, avoiding the prolonged optimization required in previous approaches. In our experiment, we introduce the Ratio Variance Warping (RVW) metric to evaluate the local geometric warping of the decal area. Extensive experimental results demonstrate that our method surpasses previous decal blending methods in terms of editing quality, editing speed and rendering speed, achieving the state-of-the-art.
Deep Rib Fracture Instance Segmentation and Classification from CT on the RibFrac Challenge
Feb 14, 2024Abstract:Rib fractures are a common and potentially severe injury that can be challenging and labor-intensive to detect in CT scans. While there have been efforts to address this field, the lack of large-scale annotated datasets and evaluation benchmarks has hindered the development and validation of deep learning algorithms. To address this issue, the RibFrac Challenge was introduced, providing a benchmark dataset of over 5,000 rib fractures from 660 CT scans, with voxel-level instance mask annotations and diagnosis labels for four clinical categories (buckle, nondisplaced, displaced, or segmental). The challenge includes two tracks: a detection (instance segmentation) track evaluated by an FROC-style metric and a classification track evaluated by an F1-style metric. During the MICCAI 2020 challenge period, 243 results were evaluated, and seven teams were invited to participate in the challenge summary. The analysis revealed that several top rib fracture detection solutions achieved performance comparable or even better than human experts. Nevertheless, the current rib fracture classification solutions are hardly clinically applicable, which can be an interesting area in the future. As an active benchmark and research resource, the data and online evaluation of the RibFrac Challenge are available at the challenge website. As an independent contribution, we have also extended our previous internal baseline by incorporating recent advancements in large-scale pretrained networks and point-based rib segmentation techniques. The resulting FracNet+ demonstrates competitive performance in rib fracture detection, which lays a foundation for further research and development in AI-assisted rib fracture detection and diagnosis.
Shifting More Attention to Breast Lesion Segmentation in Ultrasound Videos
Oct 03, 2023Abstract:Breast lesion segmentation in ultrasound (US) videos is essential for diagnosing and treating axillary lymph node metastasis. However, the lack of a well-established and large-scale ultrasound video dataset with high-quality annotations has posed a persistent challenge for the research community. To overcome this issue, we meticulously curated a US video breast lesion segmentation dataset comprising 572 videos and 34,300 annotated frames, covering a wide range of realistic clinical scenarios. Furthermore, we propose a novel frequency and localization feature aggregation network (FLA-Net) that learns temporal features from the frequency domain and predicts additional lesion location positions to assist with breast lesion segmentation. We also devise a localization-based contrastive loss to reduce the lesion location distance between neighboring video frames within the same video and enlarge the location distances between frames from different ultrasound videos. Our experiments on our annotated dataset and two public video polyp segmentation datasets demonstrate that our proposed FLA-Net achieves state-of-the-art performance in breast lesion segmentation in US videos and video polyp segmentation while significantly reducing time and space complexity. Our model and dataset are available at https://github.com/jhl-Det/FLA-Net.
Frequency-Modulated Point Cloud Rendering with Easy Editing
Mar 18, 2023



Abstract:We develop an effective point cloud rendering pipeline for novel view synthesis, which enables high fidelity local detail reconstruction, real-time rendering and user-friendly editing. In the heart of our pipeline is an adaptive frequency modulation module called Adaptive Frequency Net (AFNet), which utilizes a hypernetwork to learn the local texture frequency encoding that is consecutively injected into adaptive frequency activation layers to modulate the implicit radiance signal. This mechanism improves the frequency expressive ability of the network with richer frequency basis support, only at a small computational budget. To further boost performance, a preprocessing module is also proposed for point cloud geometry optimization via point opacity estimation. In contrast to implicit rendering, our pipeline supports high-fidelity interactive editing based on point cloud manipulation. Extensive experimental results on NeRF-Synthetic, ScanNet, DTU and Tanks and Temples datasets demonstrate the superior performances achieved by our method in terms of PSNR, SSIM and LPIPS, in comparison to the state-of-the-art.
AudioEar: Single-View Ear Reconstruction for Personalized Spatial Audio
Jan 30, 2023Abstract:Spatial audio, which focuses on immersive 3D sound rendering, is widely applied in the acoustic industry. One of the key problems of current spatial audio rendering methods is the lack of personalization based on different anatomies of individuals, which is essential to produce accurate sound source positions. In this work, we address this problem from an interdisciplinary perspective. The rendering of spatial audio is strongly correlated with the 3D shape of human bodies, particularly ears. To this end, we propose to achieve personalized spatial audio by reconstructing 3D human ears with single-view images. First, to benchmark the ear reconstruction task, we introduce AudioEar3D, a high-quality 3D ear dataset consisting of 112 point cloud ear scans with RGB images. To self-supervisedly train a reconstruction model, we further collect a 2D ear dataset composed of 2,000 images, each one with manual annotation of occlusion and 55 landmarks, named AudioEar2D. To our knowledge, both datasets have the largest scale and best quality of their kinds for public use. Further, we propose AudioEarM, a reconstruction method guided by a depth estimation network that is trained on synthetic data, with two loss functions tailored for ear data. Lastly, to fill the gap between the vision and acoustics community, we develop a pipeline to integrate the reconstructed ear mesh with an off-the-shelf 3D human body and simulate a personalized Head-Related Transfer Function (HRTF), which is the core of spatial audio rendering. Code and data are publicly available at https://github.com/seanywang0408/AudioEar.
Boosting Point Clouds Rendering via Radiance Mapping
Oct 27, 2022Abstract:Recent years we have witnessed rapid development in NeRF-based image rendering due to its high quality. However, point clouds rendering is somehow less explored. Compared to NeRF-based rendering which suffers from dense spatial sampling, point clouds rendering is naturally less computation intensive, which enables its deployment in mobile computing device. In this work, we focus on boosting the image quality of point clouds rendering with a compact model design. We first analyze the adaption of the volume rendering formulation on point clouds. Based on the analysis, we simplify the NeRF representation to a spatial mapping function which only requires single evaluation per pixel. Further, motivated by ray marching, we rectify the the noisy raw point clouds to the estimated intersection between rays and surfaces as queried coordinates, which could avoid spatial frequency collapse and neighbor point disturbance. Composed of rasterization, spatial mapping and the refinement stages, our method achieves the state-of-the-art performance on point clouds rendering, outperforming prior works by notable margins, with a smaller model size. We obtain a PSNR of 31.74 on NeRF-Synthetic, 25.88 on ScanNet and 30.81 on DTU. Code and data would be released soon.
Representation-Agnostic Shape Fields
Mar 19, 2022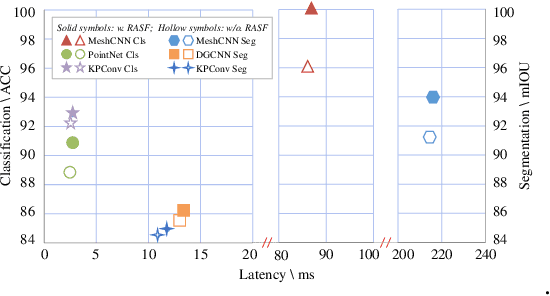
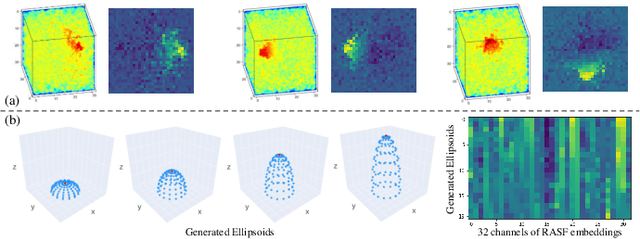
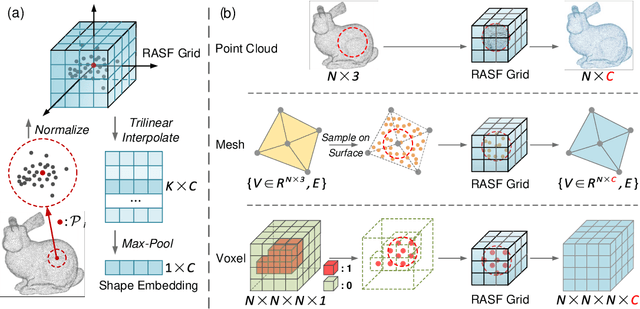
Abstract:3D shape analysis has been widely explored in the era of deep learning. Numerous models have been developed for various 3D data representation formats, e.g., MeshCNN for meshes, PointNet for point clouds and VoxNet for voxels. In this study, we present Representation-Agnostic Shape Fields (RASF), a generalizable and computation-efficient shape embedding module for 3D deep learning. RASF is implemented with a learnable 3D grid with multiple channels to store local geometry. Based on RASF, shape embeddings for various 3D shape representations (point clouds, meshes and voxels) are retrieved by coordinate indexing. While there are multiple ways to optimize the learnable parameters of RASF, we provide two effective schemes among all in this paper for RASF pre-training: shape reconstruction and normal estimation. Once trained, RASF becomes a plug-and-play performance booster with negligible cost. Extensive experiments on diverse 3D representation formats, networks and applications, validate the universal effectiveness of the proposed RASF. Code and pre-trained models are publicly available https://github.com/seanywang0408/RASF
* The Tenth International Conference on Learning Representations (ICLR 2022). Code is available at https://github.com/seanywang0408/RASF
Learning Black-Box Attackers with Transferable Priors and Query Feedback
Oct 21, 2020



Abstract:This paper addresses the challenging black-box adversarial attack problem, where only classification confidence of a victim model is available. Inspired by consistency of visual saliency between different vision models, a surrogate model is expected to improve the attack performance via transferability. By combining transferability-based and query-based black-box attack, we propose a surprisingly simple baseline approach (named SimBA++) using the surrogate model, which significantly outperforms several state-of-the-art methods. Moreover, to efficiently utilize the query feedback, we update the surrogate model in a novel learning scheme, named High-Order Gradient Approximation (HOGA). By constructing a high-order gradient computation graph, we update the surrogate model to approximate the victim model in both forward and backward pass. The SimBA++ and HOGA result in Learnable Black-Box Attack (LeBA), which surpasses previous state of the art by considerable margins: the proposed LeBA significantly reduces queries, while keeping higher attack success rates close to 100% in extensive ImageNet experiments, including attacking vision benchmarks and defensive models. Code is open source at https://github.com/TrustworthyDL/LeBA.
AlignShift: Bridging the Gap of Imaging Thickness in 3D Anisotropic Volumes
May 05, 2020



Abstract:This paper addresses a fundamental challenge in 3D medical image processing: how to deal with imaging thickness. For anisotropic medical volumes, there is a significant performance gap between thin-slice (mostly 1mm) and thick-slice (mostly 5mm) volumes. Prior arts tend to use 3D approaches for the thin-slice and 2D approaches for the thick-slice, respectively. We aim at a unified approach for both thin- and thick-slice medical volumes. Inspired by recent advances in video analysis, we propose AlignShift, a novel parameter-free operator to convert theoretically any 2D pretrained network into thickness-aware 3D network. Remarkably, the converted networks behave like 3D for the thin-slice, nevertheless degenerate to 2D for the thick-slice adaptively. The unified thickness-aware representation learning is achieved by shifting and fusing aligned "virtual slices" as per the input imaging thickness. Extensive experiments on public large-scale DeepLesion benchmark, consisting of 32K lesions for universal lesion detection, validate the effectiveness of our method, which outperforms previous state of the art by considerable margins, without whistles and bells. More importantly, to our knowledge, this is the first method that bridges the performance gap between thin- and thick-slice volumes by a unified framework. To improve research reproducibility, our code in PyTorch is open source at https://github.com/M3DV/AlignShift.
Relational Learning between Multiple Pulmonary Nodules via Deep Set Attention Transformers
Apr 12, 2020


Abstract:Diagnosis and treatment of multiple pulmonary nodules are clinically important but challenging. Prior studies on nodule characterization use solitary-nodule approaches on multiple nodular patients, which ignores the relations between nodules. In this study, we propose a multiple instance learning (MIL) approach and empirically prove the benefit to learn the relations between multiple nodules. By treating the multiple nodules from a same patient as a whole, critical relational information between solitary-nodule voxels is extracted. To our knowledge, it is the first study to learn the relations between multiple pulmonary nodules. Inspired by recent advances in natural language processing (NLP) domain, we introduce a self-attention transformer equipped with 3D CNN, named {NoduleSAT}, to replace typical pooling-based aggregation in multiple instance learning. Extensive experiments on lung nodule false positive reduction on LUNA16 database, and malignancy classification on LIDC-IDRI database, validate the effectiveness of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge