Ye Chen
ProxyImg: Towards Highly-Controllable Image Representation via Hierarchical Disentangled Proxy Embedding
Feb 02, 2026Abstract:Prevailing image representation methods, including explicit representations such as raster images and Gaussian primitives, as well as implicit representations such as latent images, either suffer from representation redundancy that leads to heavy manual editing effort, or lack a direct mapping from latent variables to semantic instances or parts, making fine-grained manipulation difficult. These limitations hinder efficient and controllable image and video editing. To address these issues, we propose a hierarchical proxy-based parametric image representation that disentangles semantic, geometric, and textural attributes into independent and manipulable parameter spaces. Based on a semantic-aware decomposition of the input image, our representation constructs hierarchical proxy geometries through adaptive Bezier fitting and iterative internal region subdivision and meshing. Multi-scale implicit texture parameters are embedded into the resulting geometry-aware distributed proxy nodes, enabling continuous high-fidelity reconstruction in the pixel domain and instance- or part-independent semantic editing. In addition, we introduce a locality-adaptive feature indexing mechanism to ensure spatial texture coherence, which further supports high-quality background completion without relying on generative models. Extensive experiments on image reconstruction and editing benchmarks, including ImageNet, OIR-Bench, and HumanEdit, demonstrate that our method achieves state-of-the-art rendering fidelity with significantly fewer parameters, while enabling intuitive, interactive, and physically plausible manipulation. Moreover, by integrating proxy nodes with Position-Based Dynamics, our framework supports real-time physics-driven animation using lightweight implicit rendering, achieving superior temporal consistency and visual realism compared with generative approaches.
Tissue Classification and Whole-Slide Images Analysis via Modeling of the Tumor Microenvironment and Biological Pathways
Jan 13, 2026Abstract:Automatic integration of whole slide images (WSIs) and gene expression profiles has demonstrated substantial potential in precision clinical diagnosis and cancer progression studies. However, most existing studies focus on individual gene sequences and slide level classification tasks, with limited attention to spatial transcriptomics and patch level applications. To address this limitation, we propose a multimodal network, BioMorphNet, which automatically integrates tissue morphological features and spatial gene expression to support tissue classification and differential gene analysis. For considering morphological features, BioMorphNet constructs a graph to model the relationships between target patches and their neighbors, and adjusts the response strength based on morphological and molecular level similarity, to better characterize the tumor microenvironment. In terms of multimodal interactions, BioMorphNet derives clinical pathway features from spatial transcriptomic data based on a predefined pathway database, serving as a bridge between tissue morphology and gene expression. In addition, a novel learnable pathway module is designed to automatically simulate the biological pathway formation process, providing a complementary representation to existing clinical pathways. Compared with the latest morphology gene multimodal methods, BioMorphNet's average classification metrics improve by 2.67%, 5.48%, and 6.29% for prostate cancer, colorectal cancer, and breast cancer datasets, respectively. BioMorphNet not only classifies tissue categories within WSIs accurately to support tumor localization, but also analyzes differential gene expression between tissue categories based on prediction confidence, contributing to the discovery of potential tumor biomarkers.
IKFST: IOO and KOO Algorithms for Accelerated and Precise WFST-based End-to-End Automatic Speech Recognition
Jan 01, 2026Abstract:End-to-end automatic speech recognition has become the dominant paradigm in both academia and industry. To enhance recognition performance, the Weighted Finite-State Transducer (WFST) is widely adopted to integrate acoustic and language models through static graph composition, providing robust decoding and effective error correction. However, WFST decoding relies on a frame-by-frame autoregressive search over CTC posterior probabilities, which severely limits inference efficiency. Motivated by establishing a more principled compatibility between WFST decoding and CTC modeling, we systematically study the two fundamental components of CTC outputs, namely blank and non-blank frames, and identify a key insight: blank frames primarily encode positional information, while non-blank frames carry semantic content. Building on this observation, we introduce Keep-Only-One and Insert-Only-One, two decoding algorithms that explicitly exploit the structural roles of blank and non-blank frames to achieve significantly faster WFST-based inference without compromising recognition accuracy. Experiments on large-scale in-house, AISHELL-1, and LibriSpeech datasets demonstrate state-of-the-art recognition accuracy with substantially reduced decoding latency, enabling truly efficient and high-performance WFST decoding in modern speech recognition systems.
3DProxyImg: Controllable 3D-Aware Animation Synthesis from Single Image via 2D-3D Aligned Proxy Embedding
Dec 19, 2025Abstract:3D animation is central to modern visual media, yet traditional production pipelines remain labor-intensive, expertise-demanding, and computationally expensive. Recent AIGC-based approaches partially automate asset creation and rigging, but they either inherit the heavy costs of full 3D pipelines or rely on video-synthesis paradigms that sacrifice 3D controllability and interactivity. We focus on single-image 3D animation generation and argue that progress is fundamentally constrained by a trade-off between rendering quality and 3D control. To address this limitation, we propose a lightweight 3D animation framework that decouples geometric control from appearance synthesis. The core idea is a 2D-3D aligned proxy representation that uses a coarse 3D estimate as a structural carrier, while delegating high-fidelity appearance and view synthesis to learned image-space generative priors. This proxy formulation enables 3D-aware motion control and interaction comparable to classical pipelines, without requiring accurate geometry or expensive optimization, and naturally extends to coherent background animation. Extensive experiments demonstrate that our method achieves efficient animation generation on low-power platforms and outperforms video-based 3D animation generation in identity preservation, geometric and textural consistency, and the level of precise, interactive control it offers to users.
Neural emulation of gravity-driven geohazard runout
Dec 18, 2025Abstract:Predicting geohazard runout is critical for protecting lives, infrastructure and ecosystems. Rapid mass flows, including landslides and avalanches, cause several thousand deaths across a wide range of environments, often travelling many kilometres from their source. The wide range of source conditions and material properties governing these flows makes their runout difficult to anticipate, particularly for downstream communities that may be suddenly exposed to severe impacts. Accurately predicting runout at scale requires models that are both physically realistic and computationally efficient, yet existing approaches face a fundamental speed-realism trade-off. Here we train a machine learning model to predict geohazard runout across representative real world terrains. The model predicts both flow extent and deposit thickness with high accuracy and 100 to 10,000 times faster computation than numerical solvers. It is trained on over 100,000 numerical simulations across over 10,000 real world digital elevation model chips and reproduces key physical behaviours, including avulsion and deposition patterns, while generalizing across different flow types, sizes and landscapes. Our results demonstrate that neural emulation enables rapid, spatially resolved runout prediction across diverse real world terrains, opening new opportunities for disaster risk reduction and impact-based forecasting. These results highlight neural emulation as a promising pathway for extending physically realistic geohazard modelling to spatial and temporal scales relevant for large scale early warning systems.
CoDA: A Context-Decoupled Hierarchical Agent with Reinforcement Learning
Dec 14, 2025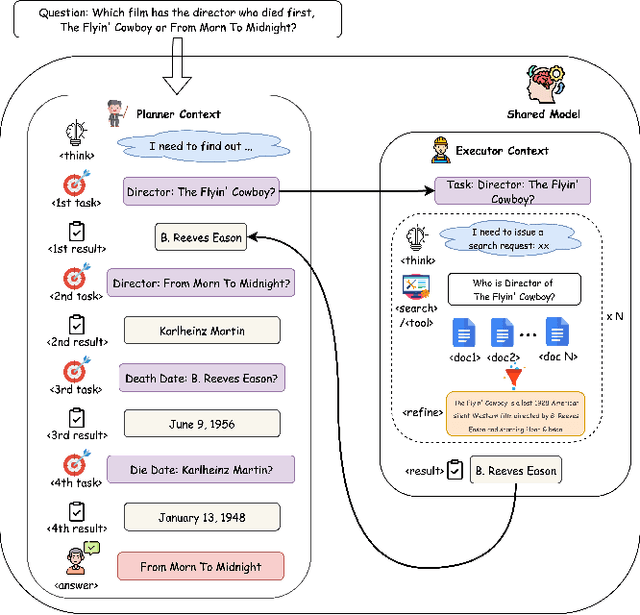
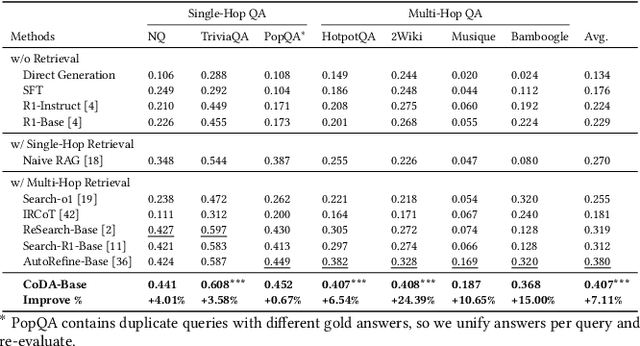
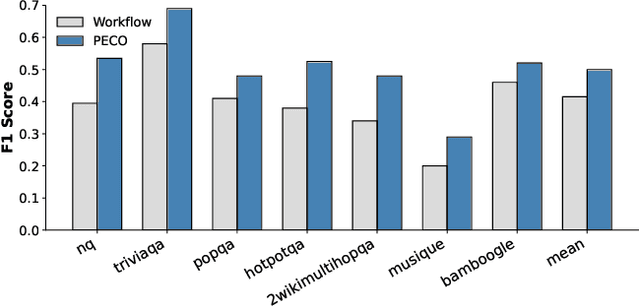
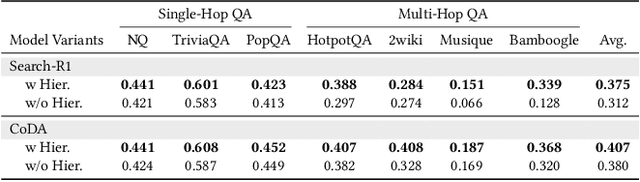
Abstract:Large Language Model (LLM) agents trained with reinforcement learning (RL) show great promise for solving complex, multi-step tasks. However, their performance is often crippled by "Context Explosion", where the accumulation of long text outputs overwhelms the model's context window and leads to reasoning failures. To address this, we introduce CoDA, a Context-Decoupled hierarchical Agent, a simple but effective reinforcement learning framework that decouples high-level planning from low-level execution. It employs a single, shared LLM backbone that learns to operate in two distinct, contextually isolated roles: a high-level Planner that decomposes tasks within a concise strategic context, and a low-level Executor that handles tool interactions in an ephemeral, isolated workspace. We train this unified agent end-to-end using PECO (Planner-Executor Co-Optimization), a reinforcement learning methodology that applies a trajectory-level reward to jointly optimize both roles, fostering seamless collaboration through context-dependent policy updates. Extensive experiments demonstrate that CoDA achieves significant performance improvements over state-of-the-art baselines on complex multi-hop question-answering benchmarks, and it exhibits strong robustness in long-context scenarios, maintaining stable performance while all other baselines suffer severe degradation, thus further validating the effectiveness of our hierarchical design in mitigating context overload.
PaintFlow: A Unified Framework for Interactive Oil Paintings Editing and Generation
Dec 09, 2025Abstract:Oil painting, as a high-level medium that blends human abstract thinking with artistic expression, poses substantial challenges for digital generation and editing due to its intricate brushstroke dynamics and stylized characteristics. Existing generation and editing techniques are often constrained by the distribution of training data and primarily focus on modifying real photographs. In this work, we introduce a unified multimodal framework for oil painting generation and editing. The proposed system allows users to incorporate reference images for precise semantic control, hand-drawn sketches for spatial structure alignment, and natural language prompts for high-level semantic guidance, while consistently maintaining a unified painting style across all outputs. Our method achieves interactive oil painting creation through three crucial technical advancements. First, we enhance the training stage with spatial alignment and semantic enhancement conditioning strategy, which map masks and sketches into spatial constraints, and encode contextual embedding from reference images and text into feature constraints, enabling object-level semantic alignment. Second, to overcome data scarcity, we propose a self-supervised style transfer pipeline based on Stroke-Based Rendering (SBR), which simulates the inpainting dynamics of oil painting restoration, converting real images into stylized oil paintings with preserved brushstroke textures to construct a large-scale paired training dataset. Finally, during inference, we integrate features using the AdaIN operator to ensure stylistic consistency. Extensive experiments demonstrate that our interactive system enables fine-grained editing while preserving the artistic qualities of oil paintings, achieving an unprecedented level of imagination realization in stylized oil paintings generation and editing.
Enhancing Agentic RL with Progressive Reward Shaping and Value-based Sampling Policy Optimization
Dec 08, 2025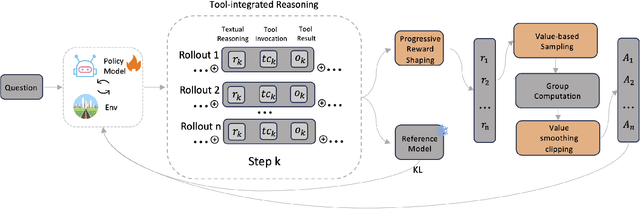
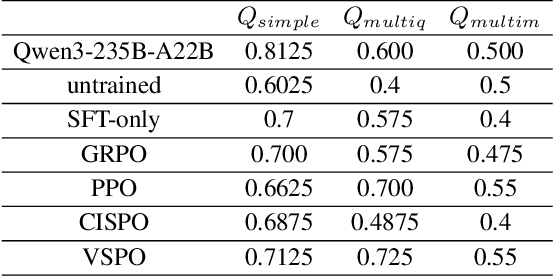
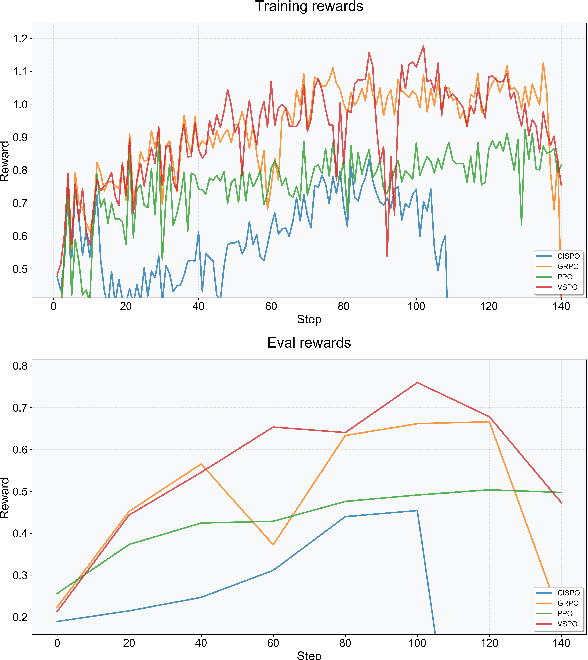
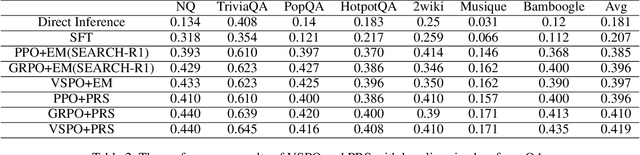
Abstract:Large Language Models (LLMs) empowered with Tool-Integrated Reasoning (TIR) can iteratively plan, call external tools, and integrate returned information to solve complex, long-horizon reasoning tasks. Agentic Reinforcement Learning (Agentic RL) optimizes such models over full tool-interaction trajectories, but two key challenges hinder effectiveness: (1) Sparse, non-instructive rewards, such as binary 0-1 verifiable signals, provide limited guidance for intermediate steps and slow convergence; (2) Gradient degradation in Group Relative Policy Optimization (GRPO), where identical rewards within a rollout group yield zero advantage, reducing sample efficiency and destabilizing training. To address these challenges, we propose two complementary techniques: Progressive Reward Shaping (PRS) and Value-based Sampling Policy Optimization (VSPO). PRS is a curriculum-inspired reward design that introduces dense, stage-wise feedback - encouraging models to first master parseable and properly formatted tool calls, then optimize for factual correctness and answer quality. We instantiate PRS for short-form QA (with a length-aware BLEU to fairly score concise answers) and long-form QA (with LLM-as-a-Judge scoring to prevent reward hacking). VSPO is an enhanced GRPO variant that replaces low-value samples with prompts selected by a task-value metric balancing difficulty and uncertainty, and applies value-smoothing clipping to stabilize gradient updates. Experiments on multiple short-form and long-form QA benchmarks show that PRS consistently outperforms traditional binary rewards, and VSPO achieves superior stability, faster convergence, and higher final performance compared to PPO, GRPO, CISPO, and SFT-only baselines. Together, PRS and VSPO yield LLM-based TIR agents that generalize better across domains.
HPR3D: Hierarchical Proxy Representation for High-Fidelity 3D Reconstruction and Controllable Editing
Jul 16, 2025



Abstract:Current 3D representations like meshes, voxels, point clouds, and NeRF-based neural implicit fields exhibit significant limitations: they are often task-specific, lacking universal applicability across reconstruction, generation, editing, and driving. While meshes offer high precision, their dense vertex data complicates editing; NeRFs deliver excellent rendering but suffer from structural ambiguity, hindering animation and manipulation; all representations inherently struggle with the trade-off between data complexity and fidelity. To overcome these issues, we introduce a novel 3D Hierarchical Proxy Node representation. Its core innovation lies in representing an object's shape and texture via a sparse set of hierarchically organized (tree-structured) proxy nodes distributed on its surface and interior. Each node stores local shape and texture information (implicitly encoded by a small MLP) within its neighborhood. Querying any 3D coordinate's properties involves efficient neural interpolation and lightweight decoding from relevant nearby and parent nodes. This framework yields a highly compact representation where nodes align with local semantics, enabling direct drag-and-edit manipulation, and offers scalable quality-complexity control. Extensive experiments across 3D reconstruction and editing demonstrate our method's expressive efficiency, high-fidelity rendering quality, and superior editability.
Long-Distance Field Demonstration of Imaging-Free Drone Identification in Intracity Environments
Apr 26, 2025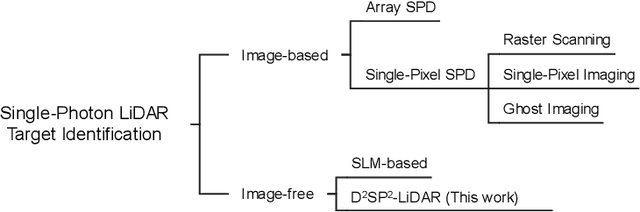
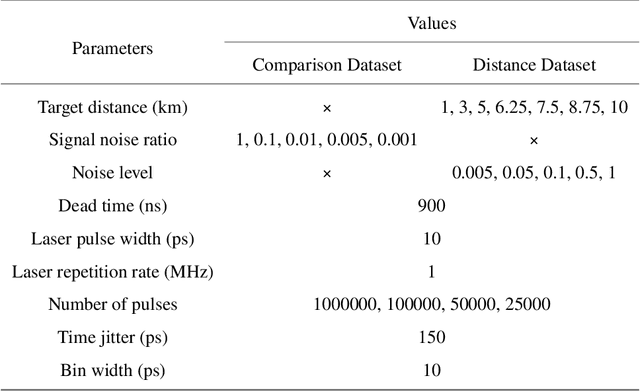
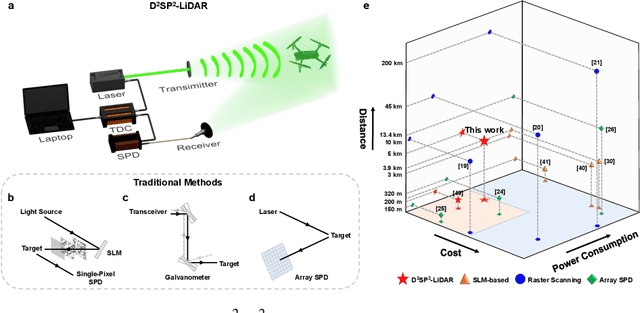
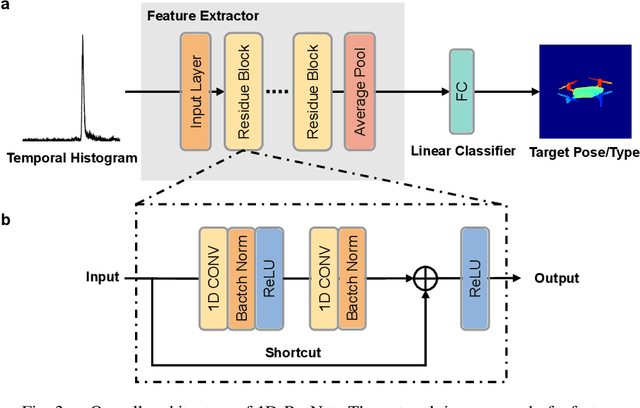
Abstract:Detecting small objects, such as drones, over long distances presents a significant challenge with broad implications for security, surveillance, environmental monitoring, and autonomous systems. Traditional imaging-based methods rely on high-resolution image acquisition, but are often constrained by range, power consumption, and cost. In contrast, data-driven single-photon-single-pixel light detection and ranging (\text{D\textsuperscript{2}SP\textsuperscript{2}-LiDAR}) provides an imaging-free alternative, directly enabling target identification while reducing system complexity and cost. However, its detection range has been limited to a few hundred meters. Here, we introduce a novel integration of residual neural networks (ResNet) with \text{D\textsuperscript{2}SP\textsuperscript{2}-LiDAR}, incorporating a refined observation model to extend the detection range to 5~\si{\kilo\meter} in an intracity environment while enabling high-accuracy identification of drone poses and types. Experimental results demonstrate that our approach not only outperforms conventional imaging-based recognition systems, but also achieves 94.93\% pose identification accuracy and 97.99\% type classification accuracy, even under weak signal conditions with long distances and low signal-to-noise ratios (SNRs). These findings highlight the potential of imaging-free methods for robust long-range detection of small targets in real-world scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge