Shixuan Gu
FreSeg: Frenet-Frame-based Part Segmentation for 3D Curvilinear Structures
Apr 19, 2024



Abstract:Part segmentation is a crucial task for 3D curvilinear structures like neuron dendrites and blood vessels, enabling the analysis of dendritic spines and aneurysms with scientific and clinical significance. However, their diversely winded morphology poses a generalization challenge to existing deep learning methods, which leads to labor-intensive manual correction. In this work, we propose FreSeg, a framework of part segmentation tasks for 3D curvilinear structures. With Frenet-Frame-based point cloud transformation, it enables the models to learn more generalizable features and have significant performance improvements on tasks involving elongated and curvy geometries. We evaluate FreSeg on 2 datasets: 1) DenSpineEM, an in-house dataset for dendritic spine segmentation, and 2) IntrA, a public 3D dataset for intracranial aneurysm segmentation. Further, we will release the DenSpineEM dataset, which includes roughly 6,000 spines from 69 dendrites from 3 public electron microscopy (EM) datasets, to foster the development of effective dendritic spine instance extraction methods and, consequently, large-scale connectivity analysis to better understand mammalian brains.
Deep Rib Fracture Instance Segmentation and Classification from CT on the RibFrac Challenge
Feb 14, 2024Abstract:Rib fractures are a common and potentially severe injury that can be challenging and labor-intensive to detect in CT scans. While there have been efforts to address this field, the lack of large-scale annotated datasets and evaluation benchmarks has hindered the development and validation of deep learning algorithms. To address this issue, the RibFrac Challenge was introduced, providing a benchmark dataset of over 5,000 rib fractures from 660 CT scans, with voxel-level instance mask annotations and diagnosis labels for four clinical categories (buckle, nondisplaced, displaced, or segmental). The challenge includes two tracks: a detection (instance segmentation) track evaluated by an FROC-style metric and a classification track evaluated by an F1-style metric. During the MICCAI 2020 challenge period, 243 results were evaluated, and seven teams were invited to participate in the challenge summary. The analysis revealed that several top rib fracture detection solutions achieved performance comparable or even better than human experts. Nevertheless, the current rib fracture classification solutions are hardly clinically applicable, which can be an interesting area in the future. As an active benchmark and research resource, the data and online evaluation of the RibFrac Challenge are available at the challenge website. As an independent contribution, we have also extended our previous internal baseline by incorporating recent advancements in large-scale pretrained networks and point-based rib segmentation techniques. The resulting FracNet+ demonstrates competitive performance in rib fracture detection, which lays a foundation for further research and development in AI-assisted rib fracture detection and diagnosis.
RibSeg v2: A Large-scale Benchmark for Rib Labeling and Anatomical Centerline Extraction
Oct 18, 2022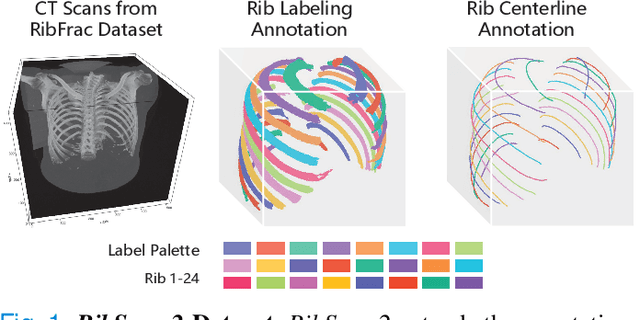
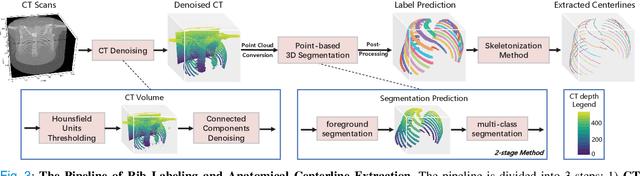
Abstract:Automatic rib labeling and anatomical centerline extraction are common prerequisites for various clinical applications. Prior studies either use in-house datasets that are inaccessible to communities, or focus on rib segmentation that neglects the clinical significance of rib labeling. To address these issues, we extend our prior dataset (RibSeg) on the binary rib segmentation task to a comprehensive benchmark, named RibSeg v2, with 660 CT scans (15,466 individual ribs in total) and annotations manually inspected by experts for rib labeling and anatomical centerline extraction. Based on the RibSeg v2, we develop a pipeline including deep learning-based methods for rib labeling, and a skeletonization-based method for centerline extraction. To improve computational efficiency, we propose a sparse point cloud representation of CT scans and compare it with standard dense voxel grids. Moreover, we design and analyze evaluation metrics to address the key challenges of each task. Our dataset, code, and model are available online to facilitate open research at https://github.com/M3DV/RibSeg
RibSeg Dataset and Strong Point Cloud Baselines for Rib Segmentation from CT Scans
Sep 17, 2021



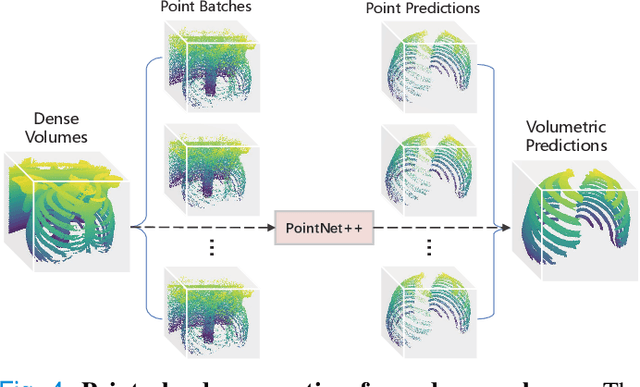
Abstract:Manual rib inspections in computed tomography (CT) scans are clinically critical but labor-intensive, as 24 ribs are typically elongated and oblique in 3D volumes. Automatic rib segmentation methods can speed up the process through rib measurement and visualization. However, prior arts mostly use in-house labeled datasets that are publicly unavailable and work on dense 3D volumes that are computationally inefficient. To address these issues, we develop a labeled rib segmentation benchmark, named \emph{RibSeg}, including 490 CT scans (11,719 individual ribs) from a public dataset. For ground truth generation, we used existing morphology-based algorithms and manually refined its results. Then, considering the sparsity of ribs in 3D volumes, we thresholded and sampled sparse voxels from the input and designed a point cloud-based baseline method for rib segmentation. The proposed method achieves state-of-the-art segmentation performance (Dice~$\approx95\%$) with significant efficiency ($10\sim40\times$ faster than prior arts). The RibSeg dataset, code, and model in PyTorch are available at https://github.com/M3DV/RibSeg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge