Jason Ken Adhinarta
FreSeg: Frenet-Frame-based Part Segmentation for 3D Curvilinear Structures
Apr 19, 2024



Abstract:Part segmentation is a crucial task for 3D curvilinear structures like neuron dendrites and blood vessels, enabling the analysis of dendritic spines and aneurysms with scientific and clinical significance. However, their diversely winded morphology poses a generalization challenge to existing deep learning methods, which leads to labor-intensive manual correction. In this work, we propose FreSeg, a framework of part segmentation tasks for 3D curvilinear structures. With Frenet-Frame-based point cloud transformation, it enables the models to learn more generalizable features and have significant performance improvements on tasks involving elongated and curvy geometries. We evaluate FreSeg on 2 datasets: 1) DenSpineEM, an in-house dataset for dendritic spine segmentation, and 2) IntrA, a public 3D dataset for intracranial aneurysm segmentation. Further, we will release the DenSpineEM dataset, which includes roughly 6,000 spines from 69 dendrites from 3 public electron microscopy (EM) datasets, to foster the development of effective dendritic spine instance extraction methods and, consequently, large-scale connectivity analysis to better understand mammalian brains.
TriSAM: Tri-Plane SAM for zero-shot cortical blood vessel segmentation in VEM images
Jan 25, 2024
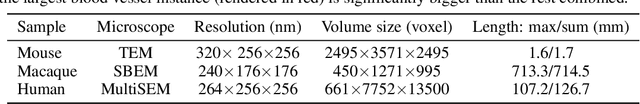

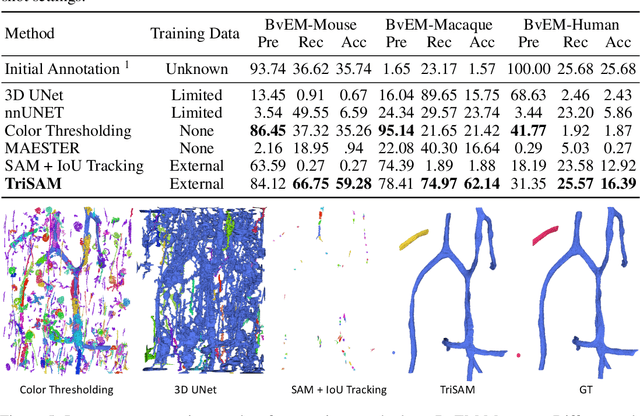
Abstract:In this paper, we address a significant gap in the field of neuroimaging by introducing the largest-to-date public benchmark, BvEM, designed specifically for cortical blood vessel segmentation in Volume Electron Microscopy (VEM) images. The intricate relationship between cerebral blood vessels and neural function underscores the vital role of vascular analysis in understanding brain health. While imaging techniques at macro and mesoscales have garnered substantial attention and resources, the microscale VEM imaging, capable of revealing intricate vascular details, has lacked the necessary benchmarking infrastructure. As researchers delve deeper into the microscale intricacies of cerebral vasculature, our BvEM benchmark represents a critical step toward unraveling the mysteries of neurovascular coupling and its impact on brain function and pathology. The BvEM dataset is based on VEM image volumes from three mammal species: adult mouse, macaque, and human. We standardized the resolution, addressed imaging variations, and meticulously annotated blood vessels through semi-automatic, manual, and quality control processes, ensuring high-quality 3D segmentation. Furthermore, we developed a zero-shot cortical blood vessel segmentation method named TriSAM, which leverages the powerful segmentation model SAM for 3D segmentation. To lift SAM from 2D segmentation to 3D volume segmentation, TriSAM employs a multi-seed tracking framework, leveraging the reliability of certain image planes for tracking while using others to identify potential turning points. This approach, consisting of Tri-Plane selection, SAM-based tracking, and recursive redirection, effectively achieves long-term 3D blood vessel segmentation without model training or fine-tuning. Experimental results show that TriSAM achieved superior performances on the BvEM benchmark across three species.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge