Jessica Zhang
FreSeg: Frenet-Frame-based Part Segmentation for 3D Curvilinear Structures
Apr 19, 2024



Abstract:Part segmentation is a crucial task for 3D curvilinear structures like neuron dendrites and blood vessels, enabling the analysis of dendritic spines and aneurysms with scientific and clinical significance. However, their diversely winded morphology poses a generalization challenge to existing deep learning methods, which leads to labor-intensive manual correction. In this work, we propose FreSeg, a framework of part segmentation tasks for 3D curvilinear structures. With Frenet-Frame-based point cloud transformation, it enables the models to learn more generalizable features and have significant performance improvements on tasks involving elongated and curvy geometries. We evaluate FreSeg on 2 datasets: 1) DenSpineEM, an in-house dataset for dendritic spine segmentation, and 2) IntrA, a public 3D dataset for intracranial aneurysm segmentation. Further, we will release the DenSpineEM dataset, which includes roughly 6,000 spines from 69 dendrites from 3 public electron microscopy (EM) datasets, to foster the development of effective dendritic spine instance extraction methods and, consequently, large-scale connectivity analysis to better understand mammalian brains.
Multimodal Dialogue Response Generation
Oct 16, 2021
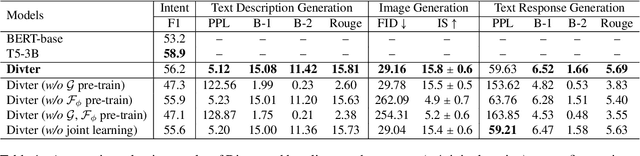
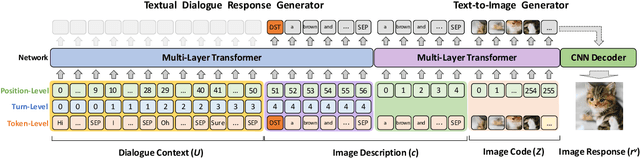
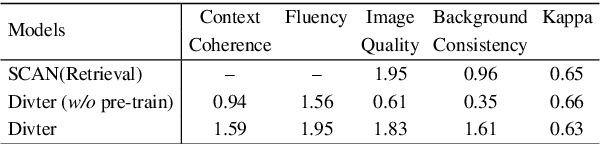
Abstract:Responsing with image has been recognized as an important capability for an intelligent conversational agent. Yet existing works only focus on exploring the multimodal dialogue models which depend on retrieval-based methods, but neglecting generation methods. To fill in the gaps, we first present a multimodal dialogue generation model, which takes the dialogue history as input, then generates a textual sequence or an image as response. Learning such a model often requires multimodal dialogues containing both texts and images which are difficult to obtain. Motivated by the challenge in practice, we consider multimodal dialogue generation under a natural assumption that only limited training examples are available. In such a low-resource setting, we devise a novel conversational agent, Divter, in order to isolate parameters that depend on multimodal dialogues from the entire generation model. By this means, the major part of the model can be learned from a large number of text-only dialogues and text-image pairs respectively, then the whole parameters can be well fitted using the limited training examples. Extensive experiments demonstrate our method achieves state-of-the-art results in both automatic and human evaluation, and can generate informative text and high-resolution image responses.
GapPredict: A Language Model for Resolving Gaps in Draft Genome Assemblies
May 25, 2021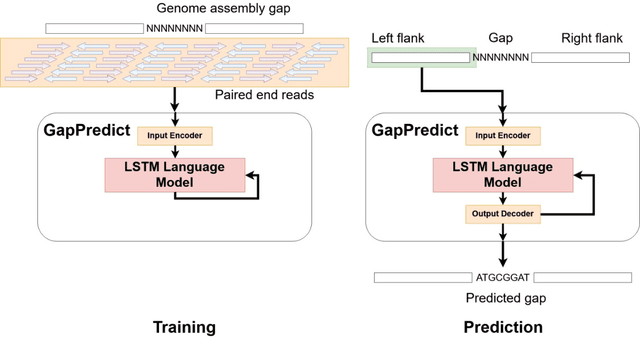
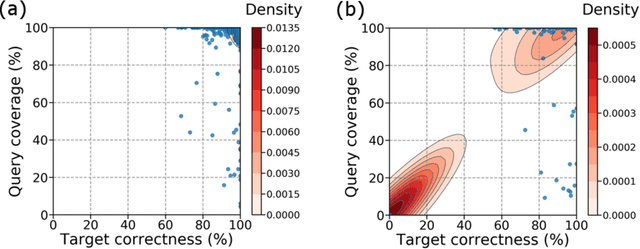
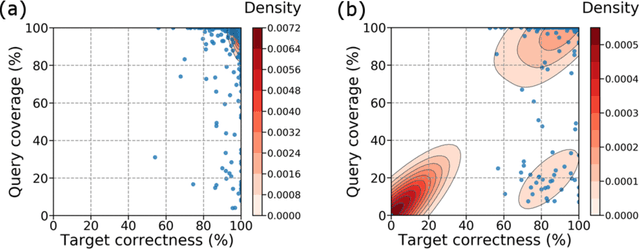
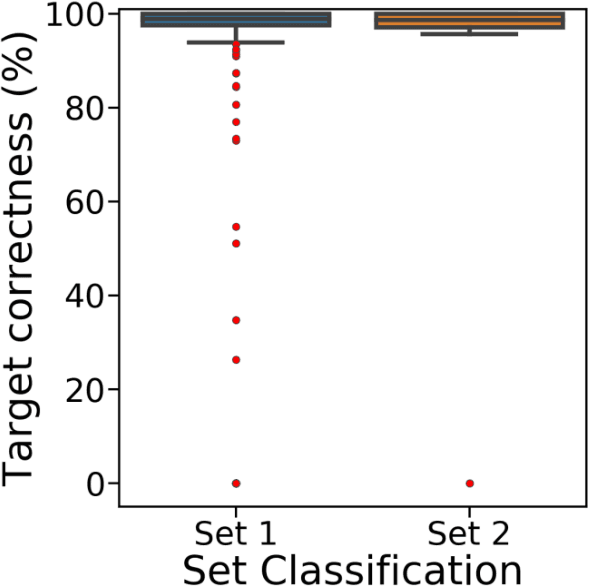
Abstract:Short-read DNA sequencing instruments can yield over 1e+12 bases per run, typically composed of reads 150 bases long. Despite this high throughput, de novo assembly algorithms have difficulty reconstructing contiguous genome sequences using short reads due to both repetitive and difficult-to-sequence regions in these genomes. Some of the short read assembly challenges are mitigated by scaffolding assembled sequences using paired-end reads. However, unresolved sequences in these scaffolds appear as "gaps". Here, we introduce GapPredict, a tool that uses a character-level language model to predict unresolved nucleotides in scaffold gaps. We benchmarked GapPredict against the state-of-the-art gap-filling tool Sealer, and observed that the former can fill 65.6% of the sampled gaps that were left unfilled by the latter, demonstrating the practical utility of deep learning approaches to the gap-filling problem in genome sequence assembly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge