Jeff Lichtman
TriSAM: Tri-Plane SAM for zero-shot cortical blood vessel segmentation in VEM images
Jan 25, 2024
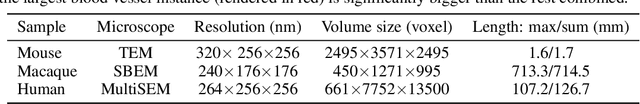

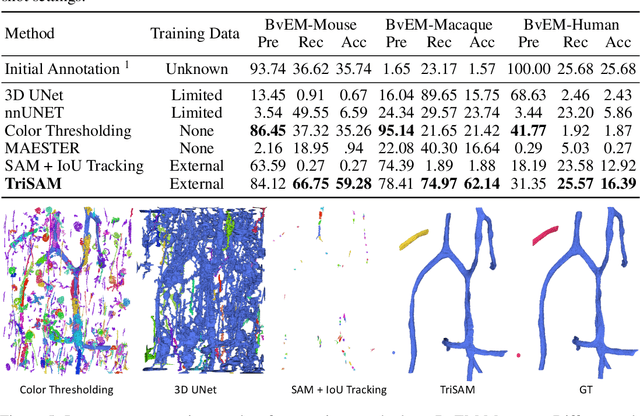
Abstract:In this paper, we address a significant gap in the field of neuroimaging by introducing the largest-to-date public benchmark, BvEM, designed specifically for cortical blood vessel segmentation in Volume Electron Microscopy (VEM) images. The intricate relationship between cerebral blood vessels and neural function underscores the vital role of vascular analysis in understanding brain health. While imaging techniques at macro and mesoscales have garnered substantial attention and resources, the microscale VEM imaging, capable of revealing intricate vascular details, has lacked the necessary benchmarking infrastructure. As researchers delve deeper into the microscale intricacies of cerebral vasculature, our BvEM benchmark represents a critical step toward unraveling the mysteries of neurovascular coupling and its impact on brain function and pathology. The BvEM dataset is based on VEM image volumes from three mammal species: adult mouse, macaque, and human. We standardized the resolution, addressed imaging variations, and meticulously annotated blood vessels through semi-automatic, manual, and quality control processes, ensuring high-quality 3D segmentation. Furthermore, we developed a zero-shot cortical blood vessel segmentation method named TriSAM, which leverages the powerful segmentation model SAM for 3D segmentation. To lift SAM from 2D segmentation to 3D volume segmentation, TriSAM employs a multi-seed tracking framework, leveraging the reliability of certain image planes for tracking while using others to identify potential turning points. This approach, consisting of Tri-Plane selection, SAM-based tracking, and recursive redirection, effectively achieves long-term 3D blood vessel segmentation without model training or fine-tuning. Experimental results show that TriSAM achieved superior performances on the BvEM benchmark across three species.
PyTorch Connectomics: A Scalable and Flexible Segmentation Framework for EM Connectomics
Dec 10, 2021



Abstract:We present PyTorch Connectomics (PyTC), an open-source deep-learning framework for the semantic and instance segmentation of volumetric microscopy images, built upon PyTorch. We demonstrate the effectiveness of PyTC in the field of connectomics, which aims to segment and reconstruct neurons, synapses, and other organelles like mitochondria at nanometer resolution for understanding neuronal communication, metabolism, and development in animal brains. PyTC is a scalable and flexible toolbox that tackles datasets at different scales and supports multi-task and semi-supervised learning to better exploit expensive expert annotations and the vast amount of unlabeled data during training. Those functionalities can be easily realized in PyTC by changing the configuration options without coding and adapted to other 2D and 3D segmentation tasks for different tissues and imaging modalities. Quantitatively, our framework achieves the best performance in the CREMI challenge for synaptic cleft segmentation (outperforms existing best result by relatively 6.1$\%$) and competitive performance on mitochondria and neuronal nuclei segmentation. Code and tutorials are publicly available at https://connectomics.readthedocs.io.
NucMM Dataset: 3D Neuronal Nuclei Instance Segmentation at Sub-Cubic Millimeter Scale
Jul 13, 2021



Abstract:Segmenting 3D cell nuclei from microscopy image volumes is critical for biological and clinical analysis, enabling the study of cellular expression patterns and cell lineages. However, current datasets for neuronal nuclei usually contain volumes smaller than $10^{\text{-}3}\ mm^3$ with fewer than 500 instances per volume, unable to reveal the complexity in large brain regions and restrict the investigation of neuronal structures. In this paper, we have pushed the task forward to the sub-cubic millimeter scale and curated the NucMM dataset with two fully annotated volumes: one $0.1\ mm^3$ electron microscopy (EM) volume containing nearly the entire zebrafish brain with around 170,000 nuclei; and one $0.25\ mm^3$ micro-CT (uCT) volume containing part of a mouse visual cortex with about 7,000 nuclei. With two imaging modalities and significantly increased volume size and instance numbers, we discover a great diversity of neuronal nuclei in appearance and density, introducing new challenges to the field. We also perform a statistical analysis to illustrate those challenges quantitatively. To tackle the challenges, we propose a novel hybrid-representation learning model that combines the merits of foreground mask, contour map, and signed distance transform to produce high-quality 3D masks. The benchmark comparisons on the NucMM dataset show that our proposed method significantly outperforms state-of-the-art nuclei segmentation approaches. Code and data are available at https://connectomics-bazaar.github.io/proj/nucMM/index.html.
AxonEM Dataset: 3D Axon Instance Segmentation of Brain Cortical Regions
Jul 12, 2021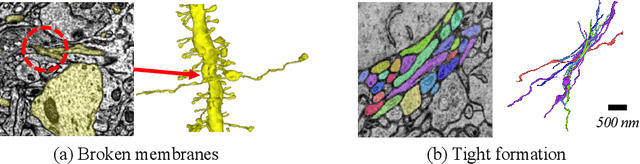

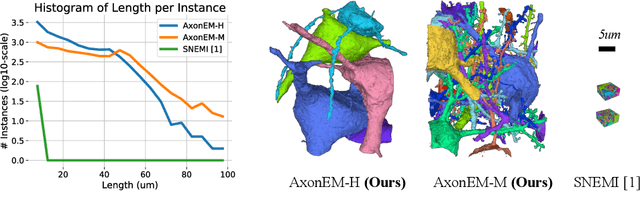
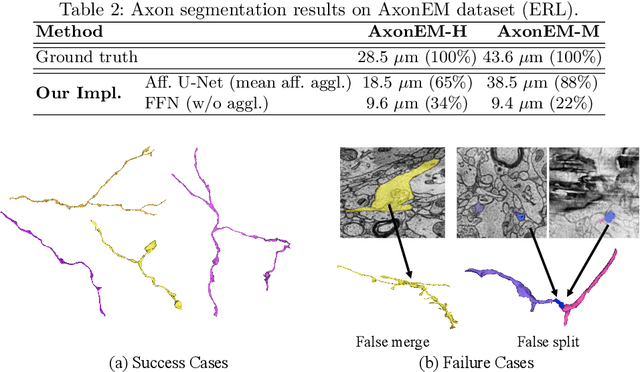
Abstract:Electron microscopy (EM) enables the reconstruction of neural circuits at the level of individual synapses, which has been transformative for scientific discoveries. However, due to the complex morphology, an accurate reconstruction of cortical axons has become a major challenge. Worse still, there is no publicly available large-scale EM dataset from the cortex that provides dense ground truth segmentation for axons, making it difficult to develop and evaluate large-scale axon reconstruction methods. To address this, we introduce the AxonEM dataset, which consists of two 30x30x30 um^3 EM image volumes from the human and mouse cortex, respectively. We thoroughly proofread over 18,000 axon instances to provide dense 3D axon instance segmentation, enabling large-scale evaluation of axon reconstruction methods. In addition, we densely annotate nine ground truth subvolumes for training, per each data volume. With this, we reproduce two published state-of-the-art methods and provide their evaluation results as a baseline. We publicly release our code and data at https://connectomics-bazaar.github.io/proj/AxonEM/index.html to foster the development of advanced methods.
Icon: An Interactive Approach to Train Deep Neural Networks for Segmentation of Neuronal Structures
Oct 27, 2016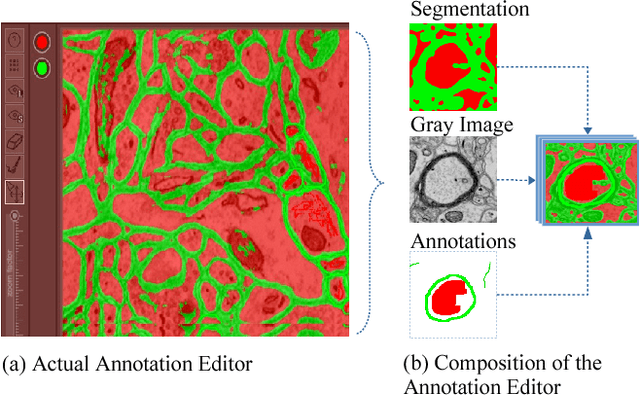
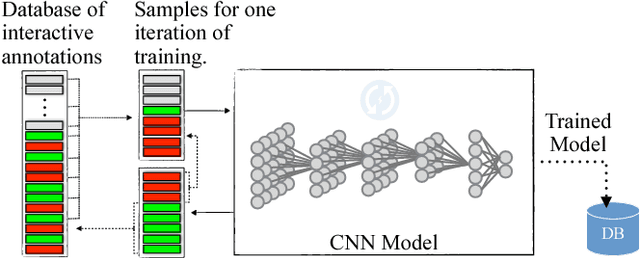
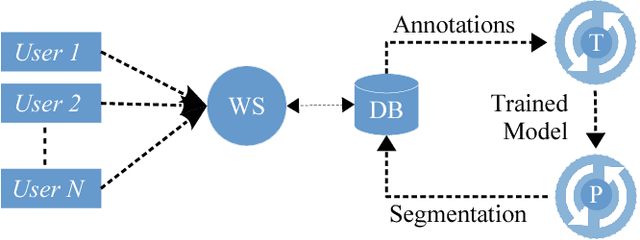
Abstract:We present an interactive approach to train a deep neural network pixel classifier for the segmentation of neuronal structures. An interactive training scheme reduces the extremely tedious manual annotation task that is typically required for deep networks to perform well on image segmentation problems. Our proposed method employs a feedback loop that captures sparse annotations using a graphical user interface, trains a deep neural network based on recent and past annotations, and displays the prediction output to users in almost real-time. Our implementation of the algorithm also allows multiple users to provide annotations in parallel and receive feedback from the same classifier. Quick feedback on classifier performance in an interactive setting enables users to identify and label examples that are more important than others for segmentation purposes. Our experiments show that an interactively-trained pixel classifier produces better region segmentation results on Electron Microscopy (EM) images than those generated by a network of the same architecture trained offline on exhaustive ground-truth labels.
Large-Scale Automatic Reconstruction of Neuronal Processes from Electron Microscopy Images
Mar 28, 2013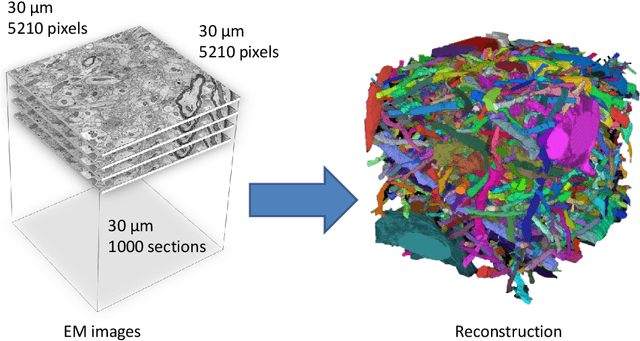
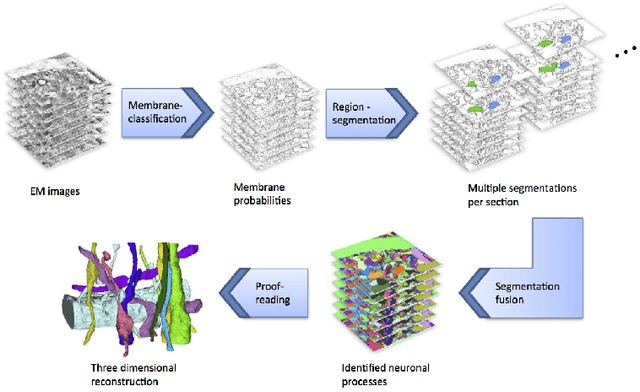
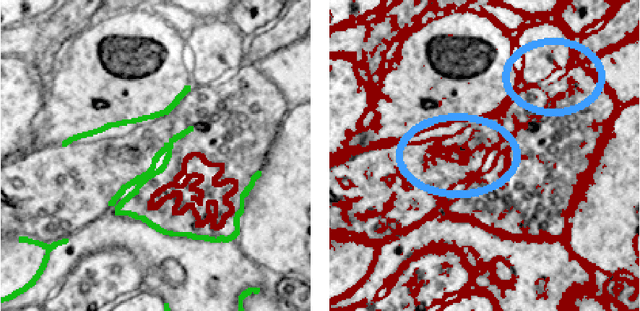
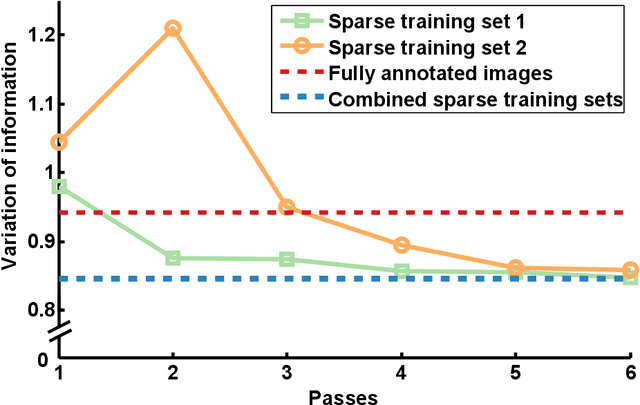
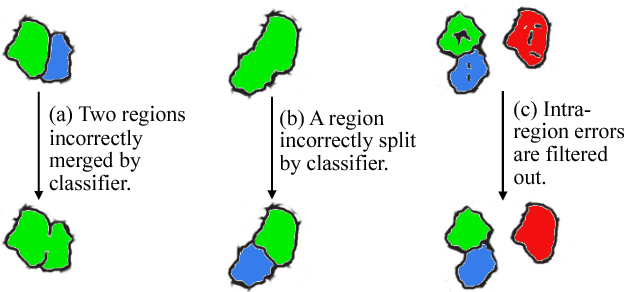
Abstract:Automated sample preparation and electron microscopy enables acquisition of very large image data sets. These technical advances are of special importance to the field of neuroanatomy, as 3D reconstructions of neuronal processes at the nm scale can provide new insight into the fine grained structure of the brain. Segmentation of large-scale electron microscopy data is the main bottleneck in the analysis of these data sets. In this paper we present a pipeline that provides state-of-the art reconstruction performance while scaling to data sets in the GB-TB range. First, we train a random forest classifier on interactive sparse user annotations. The classifier output is combined with an anisotropic smoothing prior in a Conditional Random Field framework to generate multiple segmentation hypotheses per image. These segmentations are then combined into geometrically consistent 3D objects by segmentation fusion. We provide qualitative and quantitative evaluation of the automatic segmentation and demonstrate large-scale 3D reconstructions of neuronal processes from a $\mathbf{27,000}$ $\mathbf{\mu m^3}$ volume of brain tissue over a cube of $\mathbf{30 \; \mu m}$ in each dimension corresponding to 1000 consecutive image sections. We also introduce Mojo, a proofreading tool including semi-automated correction of merge errors based on sparse user scribbles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge