Eric Miller
Discovering group dynamics in synchronous time series via hierarchical recurrent switching-state models
Jan 26, 2024Abstract:We seek to model a collection of time series arising from multiple entities interacting over the same time period. Recent work focused on modeling individual time series is inadequate for our intended applications, where collective system-level behavior influences the trajectories of individual entities. To address such problems, we present a new hierarchical switching-state model that can be trained in an unsupervised fashion to simultaneously explain both system-level and individual-level dynamics. We employ a latent system-level discrete state Markov chain that drives latent entity-level chains which in turn govern the dynamics of each observed time series. Feedback from the observations to the chains at both the entity and system levels improves flexibility via context-dependent state transitions. Our hierarchical switching recurrent dynamical models can be learned via closed-form variational coordinate ascent updates to all latent chains that scale linearly in the number of individual time series. This is asymptotically no more costly than fitting separate models for each entity. Experiments on synthetic and real datasets show that our model can produce better forecasts of future entity behavior than existing methods. Moreover, the availability of latent state chains at both the entity and system level enables interpretation of group dynamics.
Parametric Level-sets Enhanced To Improve Reconstruction (PaLEnTIR)
Apr 21, 2022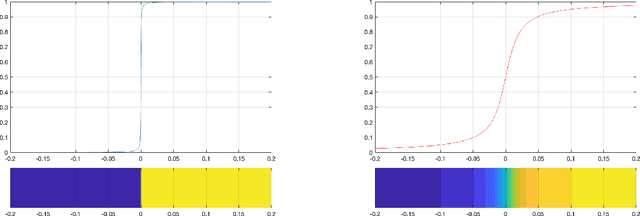
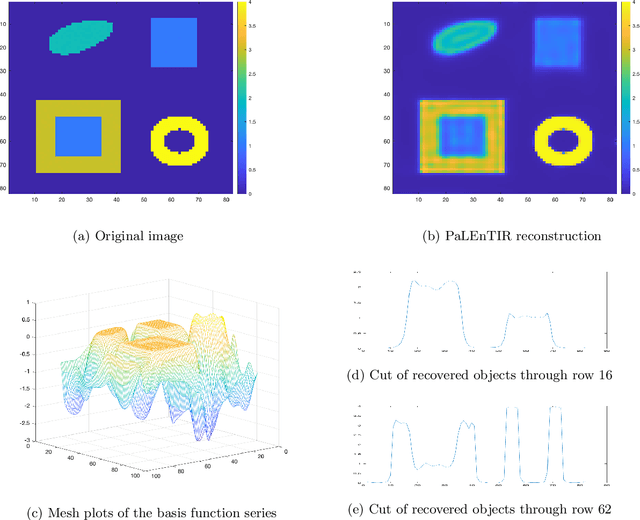
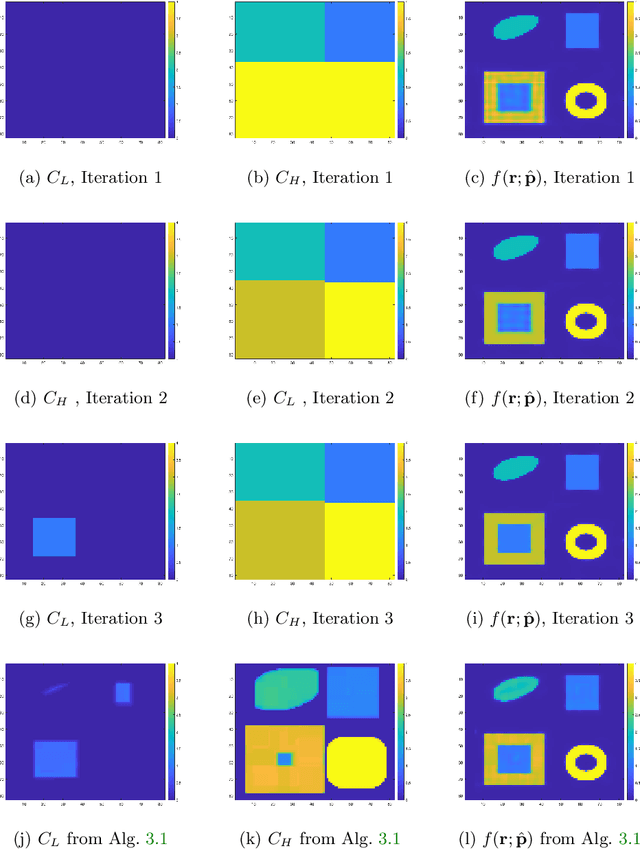

Abstract:In this paper, we consider the restoration and reconstruction of piecewise constant objects in two and three dimensions using PaLEnTIR, a significantly enhanced Parametric level set (PaLS) model relative to the current state-of-the-art. The primary contribution of this paper is a new PaLS formulation which requires only a single level set function to recover a scene with piecewise constant objects possessing multiple unknown contrasts. Our model offers distinct advantages over current approaches to the multi-contrast, multi-object problem, all of which require multiple level sets and explicit estimation of the contrast magnitudes. Given upper and lower bounds on the contrast, our approach is able to recover objects with any distribution of contrasts and eliminates the need to know either the number of contrasts in a given scene or their values. We provide an iterative process for finding these space-varying contrast limits. Relative to most PaLS methods which employ radial basis functions (RBFs), our model makes use of non-isotropic basis functions, thereby expanding the class of shapes that a PaLS model of a given complexity can approximate. Finally, PaLEnTIR improves the conditioning of the Jacobian matrix required as part of the parameter identification process and consequently accelerates the optimization methods by controlling the magnitude of the PaLS expansion coefficients, fixing the centers of the basis functions, and the uniqueness of parametric to image mappings provided by the new parameterization. We demonstrate the performance of the new approach using both 2D and 3D variants of X-ray computed tomography, diffuse optical tomography (DOT), denoising, deconvolution problems. Application to experimental sparse CT data and simulated data with different types of noise are performed to further validate the proposed method.
Interpretable contrastive word mover's embedding
Nov 01, 2021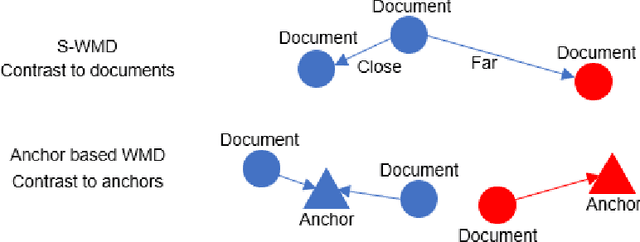
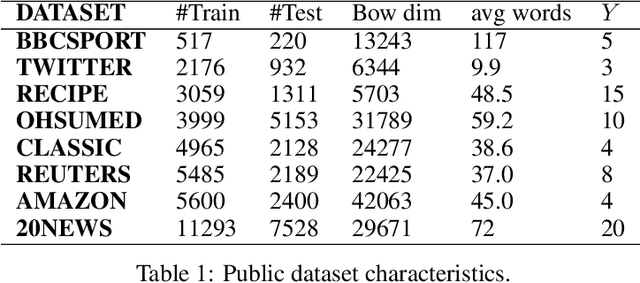
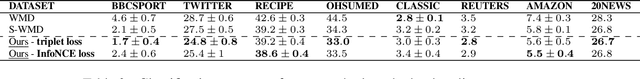
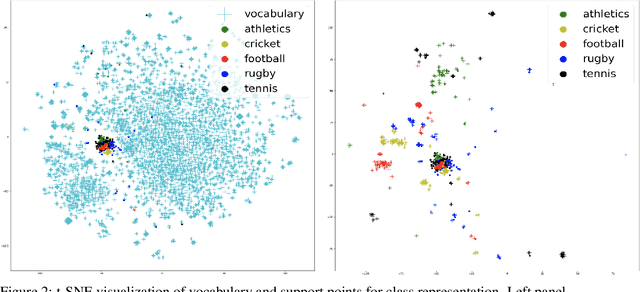
Abstract:This paper shows that a popular approach to the supervised embedding of documents for classification, namely, contrastive Word Mover's Embedding, can be significantly enhanced by adding interpretability. This interpretability is achieved by incorporating a clustering promoting mechanism into the contrastive loss. On several public datasets, we show that our method improves significantly upon existing baselines while providing interpretation to the clusters via identifying a set of keywords that are the most representative of a particular class. Our approach was motivated in part by the need to develop Natural Language Processing (NLP) methods for the \textit{novel problem of assessing student work for scientific writing and thinking} - a problem that is central to the area of (educational) Learning Sciences (LS). In this context, we show that our approach leads to a meaningful assessment of the student work related to lab reports from a biology class and can help LS researchers gain insights into student understanding and assess evidence of scientific thought processes.
Automatic coding of students' writing via Contrastive Representation Learning in the Wasserstein space
Dec 01, 2020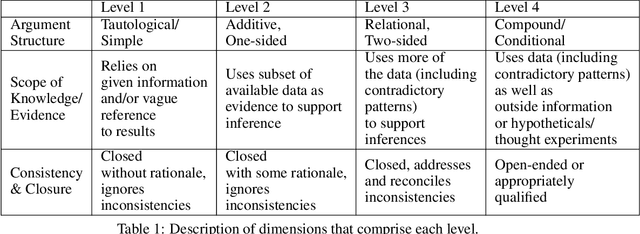
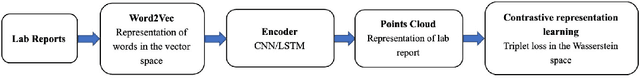
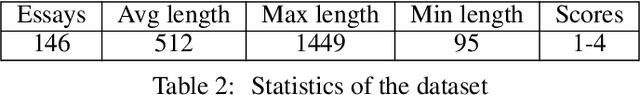
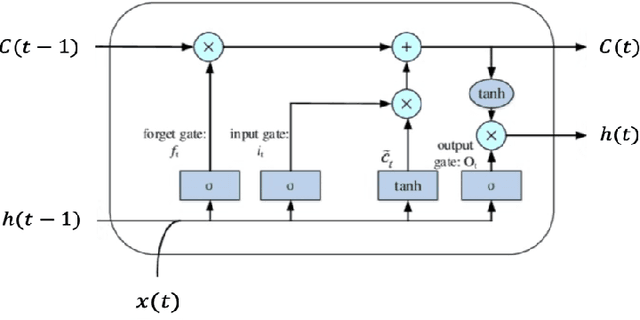
Abstract:Qualitative analysis of verbal data is of central importance in the learning sciences. It is labor-intensive and time-consuming, however, which limits the amount of data researchers can include in studies. This work is a step towards building a statistical machine learning (ML) method for achieving an automated support for qualitative analyses of students' writing, here specifically in score laboratory reports in introductory biology for sophistication of argumentation and reasoning. We start with a set of lab reports from an undergraduate biology course, scored by a four-level scheme that considers the complexity of argument structure, the scope of evidence, and the care and nuance of conclusions. Using this set of labeled data, we show that a popular natural language modeling processing pipeline, namely vector representation of words, a.k.a word embeddings, followed by Long Short Term Memory (LSTM) model for capturing language generation as a state-space model, is able to quantitatively capture the scoring, with a high Quadratic Weighted Kappa (QWK) prediction score, when trained in via a novel contrastive learning set-up. We show that the ML algorithm approached the inter-rater reliability of human analysis. Ultimately, we conclude, that machine learning (ML) for natural language processing (NLP) holds promise for assisting learning sciences researchers in conducting qualitative studies at much larger scales than is currently possible.
Large-Scale Automatic Reconstruction of Neuronal Processes from Electron Microscopy Images
Mar 28, 2013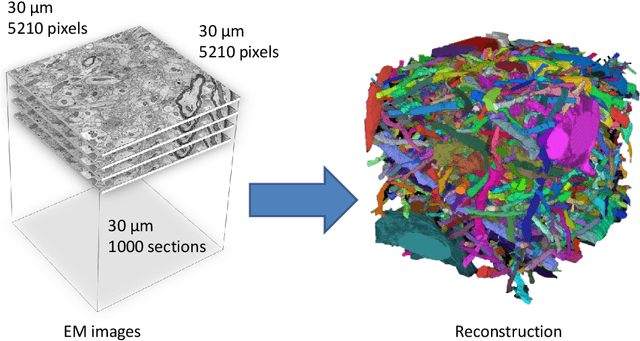
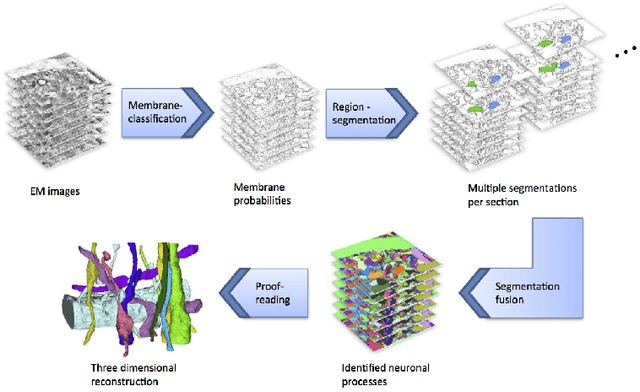
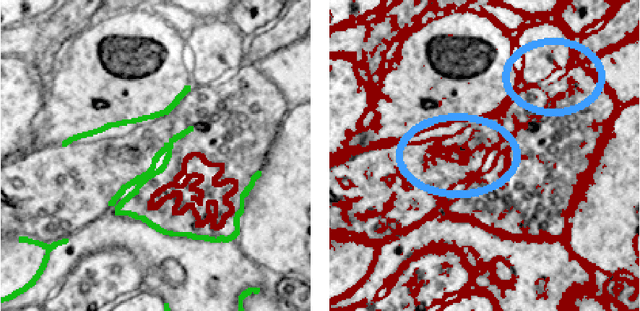
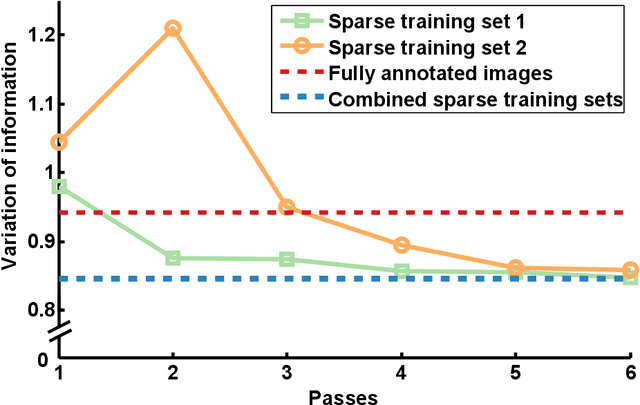
Abstract:Automated sample preparation and electron microscopy enables acquisition of very large image data sets. These technical advances are of special importance to the field of neuroanatomy, as 3D reconstructions of neuronal processes at the nm scale can provide new insight into the fine grained structure of the brain. Segmentation of large-scale electron microscopy data is the main bottleneck in the analysis of these data sets. In this paper we present a pipeline that provides state-of-the art reconstruction performance while scaling to data sets in the GB-TB range. First, we train a random forest classifier on interactive sparse user annotations. The classifier output is combined with an anisotropic smoothing prior in a Conditional Random Field framework to generate multiple segmentation hypotheses per image. These segmentations are then combined into geometrically consistent 3D objects by segmentation fusion. We provide qualitative and quantitative evaluation of the automatic segmentation and demonstrate large-scale 3D reconstructions of neuronal processes from a $\mathbf{27,000}$ $\mathbf{\mu m^3}$ volume of brain tissue over a cube of $\mathbf{30 \; \mu m}$ in each dimension corresponding to 1000 consecutive image sections. We also introduce Mojo, a proofreading tool including semi-automated correction of merge errors based on sparse user scribbles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge