Zequan Liu
PARA: Parameter-Efficient Fine-tuning with Prompt Aware Representation Adjustment
Feb 03, 2025Abstract:In the realm of parameter-efficient fine-tuning (PEFT) methods, while options like LoRA are available, there is a persistent demand in the industry for a PEFT approach that excels in both efficiency and performance within the context of single-backbone multi-tenant applications. This paper introduces a new and straightforward PEFT technique, termed \underline{P}rompt \underline{A}ware \underline{R}epresentation \underline{A}djustment (PARA). The core of our proposal is to integrate a lightweight vector generator within each Transformer layer. This generator produces vectors that are responsive to input prompts, thereby adjusting the hidden representations accordingly. Our extensive experimentation across diverse tasks has yielded promising results. Firstly, the PARA method has been shown to surpass current PEFT benchmarks in terms of performance, despite having a similar number of adjustable parameters. Secondly, it has proven to be more efficient than LoRA in the single-backbone multi-tenant scenario, highlighting its significant potential for industrial adoption.
ALoRA: Allocating Low-Rank Adaptation for Fine-tuning Large Language Models
Mar 24, 2024



Abstract:Parameter-efficient fine-tuning (PEFT) is widely studied for its effectiveness and efficiency in the era of large language models. Low-rank adaptation (LoRA) has demonstrated commendable performance as a popular and representative method. However, it is implemented with a fixed intrinsic rank that might not be the ideal setting for the downstream tasks. Recognizing the need for more flexible downstream task adaptation, we extend the methodology of LoRA to an innovative approach we call allocating low-rank adaptation (ALoRA) that enables dynamic adjustments to the intrinsic rank during the adaptation process. First, we propose a novel method, AB-LoRA, that can effectively estimate the importance score of each LoRA rank. Second, guided by AB-LoRA, we gradually prune abundant and negatively impacting LoRA ranks and allocate the pruned LoRA budgets to important Transformer modules needing higher ranks. We have conducted experiments on various tasks, and the experimental results demonstrate that our ALoRA method can outperform the recent baselines with comparable tunable parameters.
MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification
Oct 27, 2021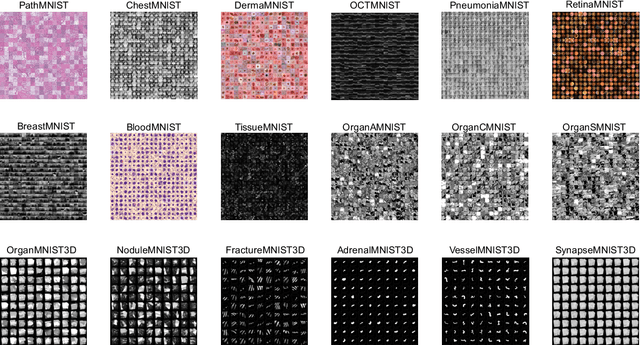
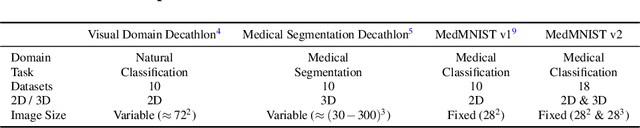
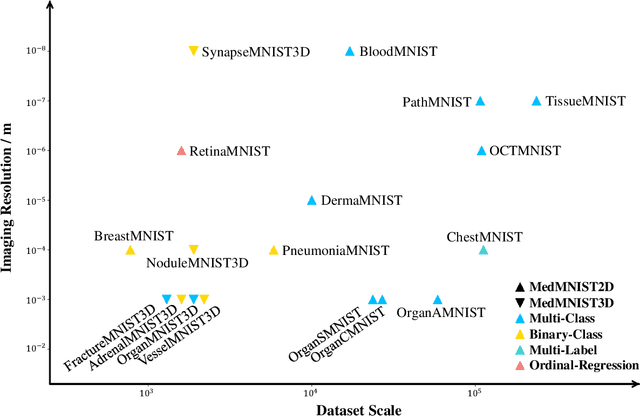
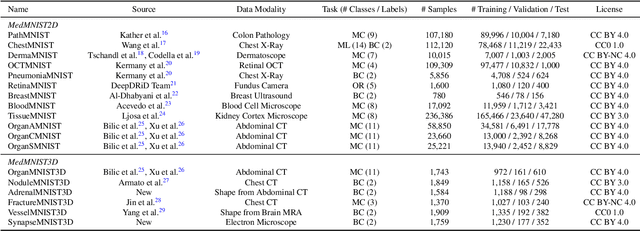
Abstract:We introduce MedMNIST v2, a large-scale MNIST-like dataset collection of standardized biomedical images, including 12 datasets for 2D and 6 datasets for 3D. All images are pre-processed into a small size of 28x28 (2D) or 28x28x28 (3D) with the corresponding classification labels so that no background knowledge is required for users. Covering primary data modalities in biomedical images, MedMNIST v2 is designed to perform classification on lightweight 2D and 3D images with various dataset scales (from 100 to 100,000) and diverse tasks (binary/multi-class, ordinal regression, and multi-label). The resulting dataset, consisting of 708,069 2D images and 10,214 3D images in total, could support numerous research / educational purposes in biomedical image analysis, computer vision, and machine learning. We benchmark several baseline methods on MedMNIST v2, including 2D / 3D neural networks and open-source / commercial AutoML tools. The data and code are publicly available at https://medmnist.com/.
AxonEM Dataset: 3D Axon Instance Segmentation of Brain Cortical Regions
Jul 12, 2021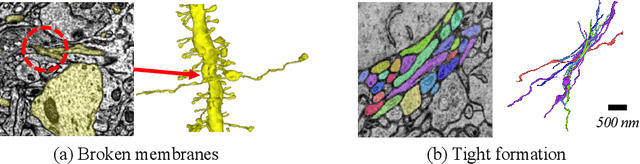

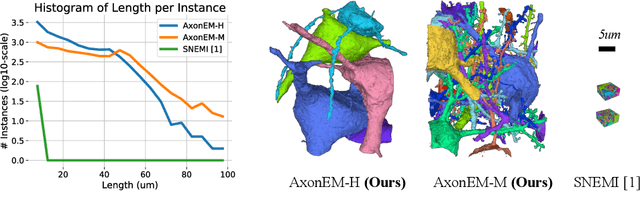
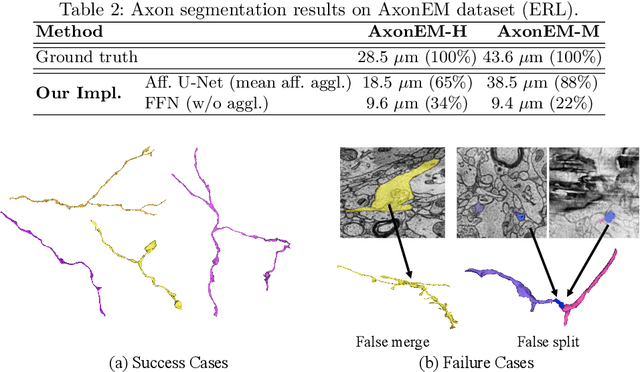
Abstract:Electron microscopy (EM) enables the reconstruction of neural circuits at the level of individual synapses, which has been transformative for scientific discoveries. However, due to the complex morphology, an accurate reconstruction of cortical axons has become a major challenge. Worse still, there is no publicly available large-scale EM dataset from the cortex that provides dense ground truth segmentation for axons, making it difficult to develop and evaluate large-scale axon reconstruction methods. To address this, we introduce the AxonEM dataset, which consists of two 30x30x30 um^3 EM image volumes from the human and mouse cortex, respectively. We thoroughly proofread over 18,000 axon instances to provide dense 3D axon instance segmentation, enabling large-scale evaluation of axon reconstruction methods. In addition, we densely annotate nine ground truth subvolumes for training, per each data volume. With this, we reproduce two published state-of-the-art methods and provide their evaluation results as a baseline. We publicly release our code and data at https://connectomics-bazaar.github.io/proj/AxonEM/index.html to foster the development of advanced methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge