Zudi Lin
Structure-Preserving Instance Segmentation via Skeleton-Aware Distance Transform
Oct 08, 2023Abstract:Objects with complex structures pose significant challenges to existing instance segmentation methods that rely on boundary or affinity maps, which are vulnerable to small errors around contacting pixels that cause noticeable connectivity change. While the distance transform (DT) makes instance interiors and boundaries more distinguishable, it tends to overlook the intra-object connectivity for instances with varying width and result in over-segmentation. To address these challenges, we propose a skeleton-aware distance transform (SDT) that combines the merits of object skeleton in preserving connectivity and DT in modeling geometric arrangement to represent instances with arbitrary structures. Comprehensive experiments on histopathology image segmentation demonstrate that SDT achieves state-of-the-art performance.
Domain-Scalable Unpaired Image Translation via Latent Space Anchoring
Jun 26, 2023



Abstract:Unpaired image-to-image translation (UNIT) aims to map images between two visual domains without paired training data. However, given a UNIT model trained on certain domains, it is difficult for current methods to incorporate new domains because they often need to train the full model on both existing and new domains. To address this problem, we propose a new domain-scalable UNIT method, termed as latent space anchoring, which can be efficiently extended to new visual domains and does not need to fine-tune encoders and decoders of existing domains. Our method anchors images of different domains to the same latent space of frozen GANs by learning lightweight encoder and regressor models to reconstruct single-domain images. In the inference phase, the learned encoders and decoders of different domains can be arbitrarily combined to translate images between any two domains without fine-tuning. Experiments on various datasets show that the proposed method achieves superior performance on both standard and domain-scalable UNIT tasks in comparison with the state-of-the-art methods.
CDDFuse: Correlation-Driven Dual-Branch Feature Decomposition for Multi-Modality Image Fusion
Nov 26, 2022



Abstract:Multi-modality (MM) image fusion aims to render fused images that maintain the merits of different modalities, e.g., functional highlight and detailed textures. To tackle the challenge in modeling cross-modality features and decomposing desirable modality-specific and modality-shared features, we propose a novel Correlation-Driven feature Decomposition Fusion (CDDFuse) network for end-to-end MM feature decomposition and image fusion. In the first stage of the two-stage architectures, CDDFuse uses Restormer blocks to extract cross-modality shallow features. We then introduce a dual-branch Transformer-CNN feature extractor with Lite Transformer (LT) blocks leveraging long-range attention to handle low-frequency global features and Invertible Neural Networks (INN) blocks focusing on extracting high-frequency local information. Upon the embedded semantic information, the low-frequency features should be correlated while the high-frequency features should be uncorrelated. Thus, we propose a correlation-driven loss for better feature decomposition. In the second stage, the LT-based global fusion and INN-based local fusion layers output the fused image. Extensive experiments demonstrate that our CDDFuse achieves promising results in multiple fusion tasks, including infrared-visible image fusion and medical image fusion. We also show that CDDFuse can boost the performance in downstream infrared-visible semantic segmentation and object detection in a unified benchmark.
MST++: Multi-stage Spectral-wise Transformer for Efficient Spectral Reconstruction
Apr 17, 2022



Abstract:Existing leading methods for spectral reconstruction (SR) focus on designing deeper or wider convolutional neural networks (CNNs) to learn the end-to-end mapping from the RGB image to its hyperspectral image (HSI). These CNN-based methods achieve impressive restoration performance while showing limitations in capturing the long-range dependencies and self-similarity prior. To cope with this problem, we propose a novel Transformer-based method, Multi-stage Spectral-wise Transformer (MST++), for efficient spectral reconstruction. In particular, we employ Spectral-wise Multi-head Self-attention (S-MSA) that is based on the HSI spatially sparse while spectrally self-similar nature to compose the basic unit, Spectral-wise Attention Block (SAB). Then SABs build up Single-stage Spectral-wise Transformer (SST) that exploits a U-shaped structure to extract multi-resolution contextual information. Finally, our MST++, cascaded by several SSTs, progressively improves the reconstruction quality from coarse to fine. Comprehensive experiments show that our MST++ significantly outperforms other state-of-the-art methods. In the NTIRE 2022 Spectral Reconstruction Challenge, our approach won the First place. Code and pre-trained models are publicly available at https://github.com/caiyuanhao1998/MST-plus-plus.
* Winner of NTIRE 2022 Challenge on Spectral Reconstruction from RGB; The First Transformer-based Method for Spectral Reconstruction
Instance Segmentation of Unlabeled Modalities via Cyclic Segmentation GAN
Apr 06, 2022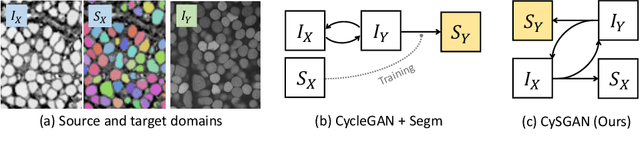
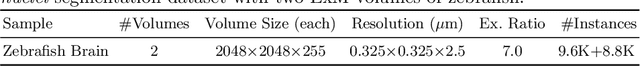
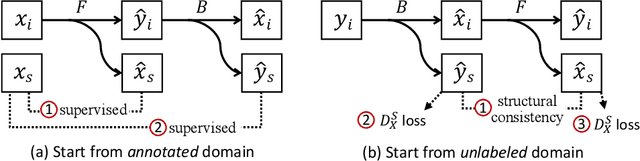
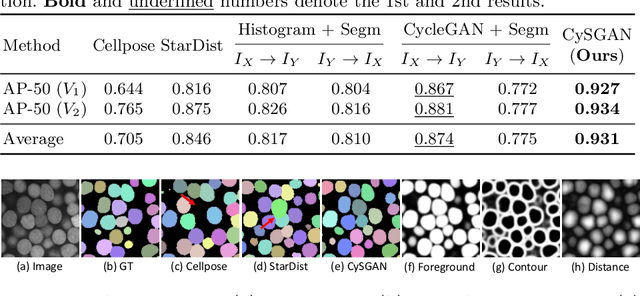
Abstract:Instance segmentation for unlabeled imaging modalities is a challenging but essential task as collecting expert annotation can be expensive and time-consuming. Existing works segment a new modality by either deploying a pre-trained model optimized on diverse training data or conducting domain translation and image segmentation as two independent steps. In this work, we propose a novel Cyclic Segmentation Generative Adversarial Network (CySGAN) that conducts image translation and instance segmentation jointly using a unified framework. Besides the CycleGAN losses for image translation and supervised losses for the annotated source domain, we introduce additional self-supervised and segmentation-based adversarial objectives to improve the model performance by leveraging unlabeled target domain images. We benchmark our approach on the task of 3D neuronal nuclei segmentation with annotated electron microscopy (EM) images and unlabeled expansion microscopy (ExM) data. Our CySGAN outperforms both pretrained generalist models and the baselines that sequentially conduct image translation and segmentation. Our implementation and the newly collected, densely annotated ExM nuclei dataset, named NucExM, are available at https://connectomics-bazaar.github.io/proj/CySGAN/index.html.
Revisiting RCAN: Improved Training for Image Super-Resolution
Jan 27, 2022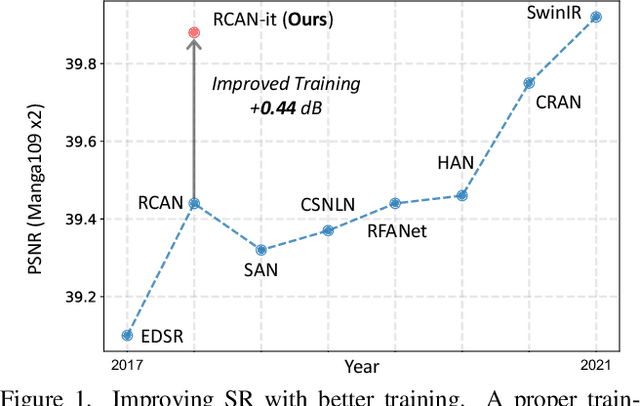
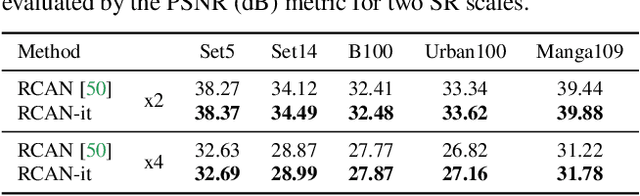
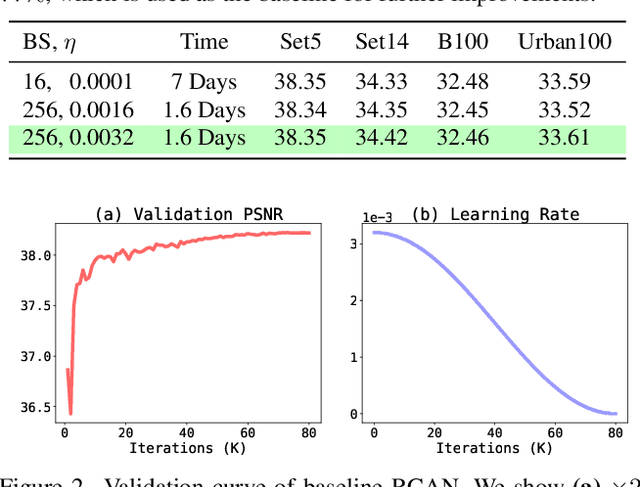
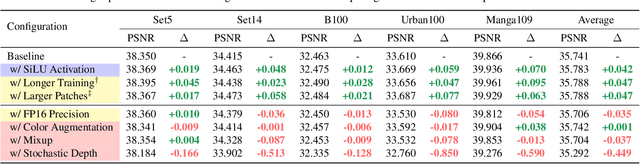
Abstract:Image super-resolution (SR) is a fast-moving field with novel architectures attracting the spotlight. However, most SR models were optimized with dated training strategies. In this work, we revisit the popular RCAN model and examine the effect of different training options in SR. Surprisingly (or perhaps as expected), we show that RCAN can outperform or match nearly all the CNN-based SR architectures published after RCAN on standard benchmarks with a proper training strategy and minimal architecture change. Besides, although RCAN is a very large SR architecture with more than four hundred convolutional layers, we draw a notable conclusion that underfitting is still the main problem restricting the model capability instead of overfitting. We observe supportive evidence that increasing training iterations clearly improves the model performance while applying regularization techniques generally degrades the predictions. We denote our simply revised RCAN as RCAN-it and recommend practitioners to use it as baselines for future research. Code is publicly available at https://github.com/zudi-lin/rcan-it.
PyTorch Connectomics: A Scalable and Flexible Segmentation Framework for EM Connectomics
Dec 10, 2021



Abstract:We present PyTorch Connectomics (PyTC), an open-source deep-learning framework for the semantic and instance segmentation of volumetric microscopy images, built upon PyTorch. We demonstrate the effectiveness of PyTC in the field of connectomics, which aims to segment and reconstruct neurons, synapses, and other organelles like mitochondria at nanometer resolution for understanding neuronal communication, metabolism, and development in animal brains. PyTC is a scalable and flexible toolbox that tackles datasets at different scales and supports multi-task and semi-supervised learning to better exploit expensive expert annotations and the vast amount of unlabeled data during training. Those functionalities can be easily realized in PyTC by changing the configuration options without coding and adapted to other 2D and 3D segmentation tasks for different tissues and imaging modalities. Quantitatively, our framework achieves the best performance in the CREMI challenge for synaptic cleft segmentation (outperforms existing best result by relatively 6.1$\%$) and competitive performance on mitochondria and neuronal nuclei segmentation. Code and tutorials are publicly available at https://connectomics.readthedocs.io.
Object Propagation via Inter-Frame Attentions for Temporally Stable Video Instance Segmentation
Nov 15, 2021



Abstract:Video instance segmentation aims to detect, segment, and track objects in a video. Current approaches extend image-level segmentation algorithms to the temporal domain. However, this results in temporally inconsistent masks. In this work, we identify the mask quality due to temporal stability as a performance bottleneck. Motivated by this, we propose a video instance segmentation method that alleviates the problem due to missing detections. Since this cannot be solved simply using spatial information, we leverage temporal context using inter-frame attentions. This allows our network to refocus on missing objects using box predictions from the neighbouring frame, thereby overcoming missing detections. Our method significantly outperforms previous state-of-the-art algorithms using the Mask R-CNN backbone, by achieving 35.1% mAP on the YouTube-VIS benchmark. Additionally, our method is completely online and requires no future frames. Our code is publicly available at https://github.com/anirudh-chakravarthy/ObjProp.
Asymmetric 3D Context Fusion for Universal Lesion Detection
Sep 17, 2021



Abstract:Modeling 3D context is essential for high-performance 3D medical image analysis. Although 2D networks benefit from large-scale 2D supervised pretraining, it is weak in capturing 3D context. 3D networks are strong in 3D context yet lack supervised pretraining. As an emerging technique, \emph{3D context fusion operator}, which enables conversion from 2D pretrained networks, leverages the advantages of both and has achieved great success. Existing 3D context fusion operators are designed to be spatially symmetric, i.e., performing identical operations on each 2D slice like convolutions. However, these operators are not truly equivariant to translation, especially when only a few 3D slices are used as inputs. In this paper, we propose a novel asymmetric 3D context fusion operator (A3D), which uses different weights to fuse 3D context from different 2D slices. Notably, A3D is NOT translation-equivariant while it significantly outperforms existing symmetric context fusion operators without introducing large computational overhead. We validate the effectiveness of the proposed method by extensive experiments on DeepLesion benchmark, a large-scale public dataset for universal lesion detection from computed tomography (CT). The proposed A3D consistently outperforms symmetric context fusion operators by considerable margins, and establishes a new \emph{state of the art} on DeepLesion. To facilitate open research, our code and model in PyTorch are available at https://github.com/M3DV/AlignShift.
NucMM Dataset: 3D Neuronal Nuclei Instance Segmentation at Sub-Cubic Millimeter Scale
Jul 13, 2021



Abstract:Segmenting 3D cell nuclei from microscopy image volumes is critical for biological and clinical analysis, enabling the study of cellular expression patterns and cell lineages. However, current datasets for neuronal nuclei usually contain volumes smaller than $10^{\text{-}3}\ mm^3$ with fewer than 500 instances per volume, unable to reveal the complexity in large brain regions and restrict the investigation of neuronal structures. In this paper, we have pushed the task forward to the sub-cubic millimeter scale and curated the NucMM dataset with two fully annotated volumes: one $0.1\ mm^3$ electron microscopy (EM) volume containing nearly the entire zebrafish brain with around 170,000 nuclei; and one $0.25\ mm^3$ micro-CT (uCT) volume containing part of a mouse visual cortex with about 7,000 nuclei. With two imaging modalities and significantly increased volume size and instance numbers, we discover a great diversity of neuronal nuclei in appearance and density, introducing new challenges to the field. We also perform a statistical analysis to illustrate those challenges quantitatively. To tackle the challenges, we propose a novel hybrid-representation learning model that combines the merits of foreground mask, contour map, and signed distance transform to produce high-quality 3D masks. The benchmark comparisons on the NucMM dataset show that our proposed method significantly outperforms state-of-the-art nuclei segmentation approaches. Code and data are available at https://connectomics-bazaar.github.io/proj/nucMM/index.html.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge