Edward S. Boyden
Real-time Neuron Segmentation for Voltage Imaging
Mar 25, 2024Abstract:In voltage imaging, where the membrane potentials of individual neurons are recorded at from hundreds to thousand frames per second using fluorescence microscopy, data processing presents a challenge. Even a fraction of a minute of recording with a limited image size yields gigabytes of video data consisting of tens of thousands of frames, which can be time-consuming to process. Moreover, millisecond-level short exposures lead to noisy video frames, obscuring neuron footprints especially in deep-brain samples where noisy signals are buried in background fluorescence. To address this challenge, we propose a fast neuron segmentation method able to detect multiple, potentially overlapping, spiking neurons from noisy video frames, and implement a data processing pipeline incorporating the proposed segmentation method along with GPU-accelerated motion correction. By testing on existing datasets as well as on new datasets we introduce, we show that our pipeline extracts neuron footprints that agree well with human annotation even from cluttered datasets, and demonstrate real-time processing of voltage imaging data on a single desktop computer for the first time.
Fast light-field 3D microscopy with out-of-distribution detection and adaptation through Conditional Normalizing Flows
Jun 14, 2023Abstract:Real-time 3D fluorescence microscopy is crucial for the spatiotemporal analysis of live organisms, such as neural activity monitoring. The eXtended field-of-view light field microscope (XLFM), also known as Fourier light field microscope, is a straightforward, single snapshot solution to achieve this. The XLFM acquires spatial-angular information in a single camera exposure. In a subsequent step, a 3D volume can be algorithmically reconstructed, making it exceptionally well-suited for real-time 3D acquisition and potential analysis. Unfortunately, traditional reconstruction methods (like deconvolution) require lengthy processing times (0.0220 Hz), hampering the speed advantages of the XLFM. Neural network architectures can overcome the speed constraints at the expense of lacking certainty metrics, which renders them untrustworthy for the biomedical realm. This work proposes a novel architecture to perform fast 3D reconstructions of live immobilized zebrafish neural activity based on a conditional normalizing flow. It reconstructs volumes at 8 Hz spanning 512x512x96 voxels, and it can be trained in under two hours due to the small dataset requirements (10 image-volume pairs). Furthermore, normalizing flows allow for exact Likelihood computation, enabling distribution monitoring, followed by out-of-distribution detection and retraining of the system when a novel sample is detected. We evaluate the proposed method on a cross-validation approach involving multiple in-distribution samples (genetically identical zebrafish) and various out-of-distribution ones.
Instance Segmentation of Unlabeled Modalities via Cyclic Segmentation GAN
Apr 06, 2022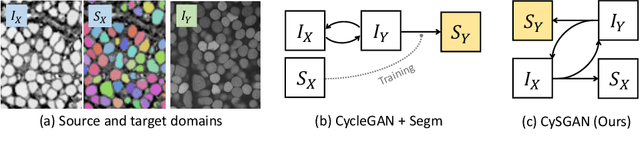
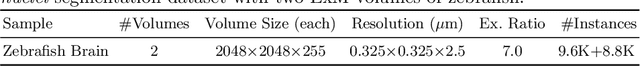
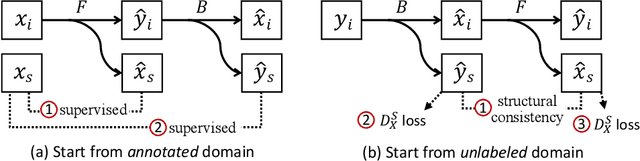
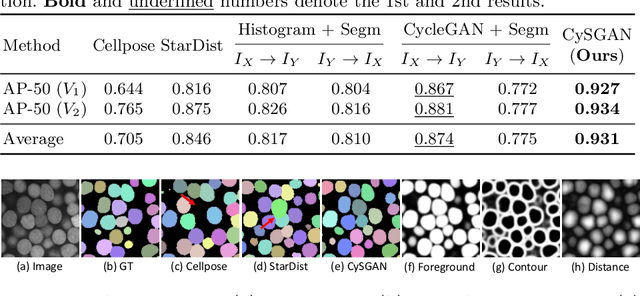
Abstract:Instance segmentation for unlabeled imaging modalities is a challenging but essential task as collecting expert annotation can be expensive and time-consuming. Existing works segment a new modality by either deploying a pre-trained model optimized on diverse training data or conducting domain translation and image segmentation as two independent steps. In this work, we propose a novel Cyclic Segmentation Generative Adversarial Network (CySGAN) that conducts image translation and instance segmentation jointly using a unified framework. Besides the CycleGAN losses for image translation and supervised losses for the annotated source domain, we introduce additional self-supervised and segmentation-based adversarial objectives to improve the model performance by leveraging unlabeled target domain images. We benchmark our approach on the task of 3D neuronal nuclei segmentation with annotated electron microscopy (EM) images and unlabeled expansion microscopy (ExM) data. Our CySGAN outperforms both pretrained generalist models and the baselines that sequentially conduct image translation and segmentation. Our implementation and the newly collected, densely annotated ExM nuclei dataset, named NucExM, are available at https://connectomics-bazaar.github.io/proj/CySGAN/index.html.
Morphological Error Detection in 3D Segmentations
May 30, 2017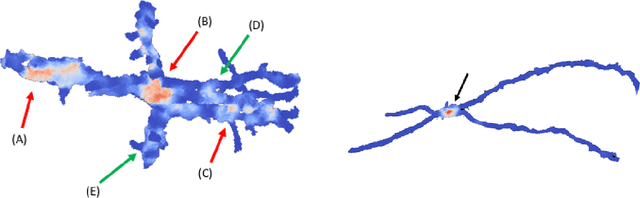
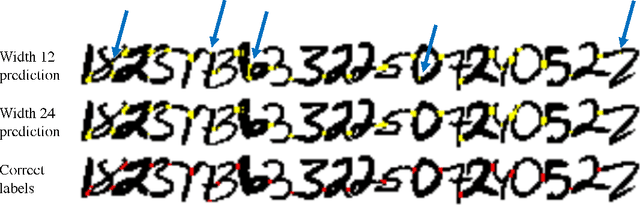
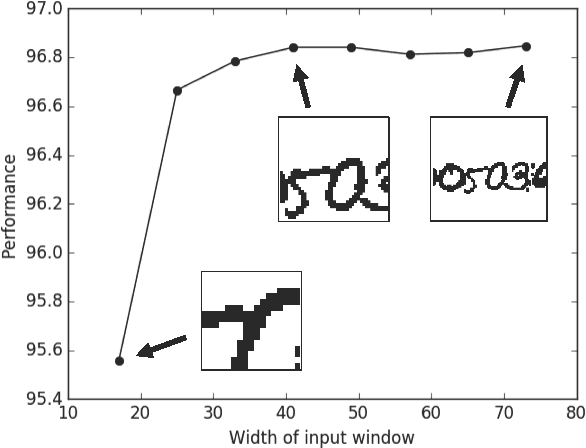
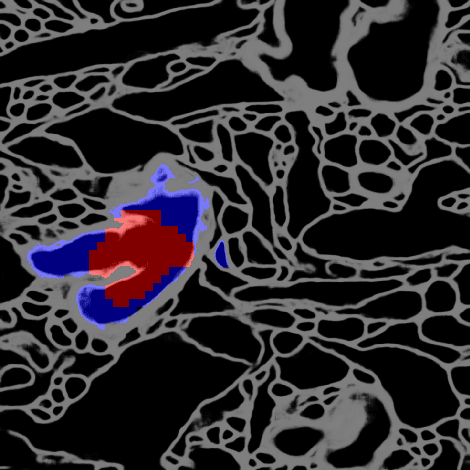
Abstract:Deep learning algorithms for connectomics rely upon localized classification, rather than overall morphology. This leads to a high incidence of erroneously merged objects. Humans, by contrast, can easily detect such errors by acquiring intuition for the correct morphology of objects. Biological neurons have complicated and variable shapes, which are challenging to learn, and merge errors take a multitude of different forms. We present an algorithm, MergeNet, that shows 3D ConvNets can, in fact, detect merge errors from high-level neuronal morphology. MergeNet follows unsupervised training and operates across datasets. We demonstrate the performance of MergeNet both on a variety of connectomics data and on a dataset created from merged MNIST images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge