Tobias Lasser
Munich Institute of Biomedical Engineering, Technical University of Munich, Germany, Computational Imaging and Inverse Problems, Department of Informatics, School of Computation, Information, and Technology, Technical University of Munich, Germany
Cosmos-Transfer1: Conditional World Generation with Adaptive Multimodal Control
Mar 18, 2025Abstract:We introduce Cosmos-Transfer, a conditional world generation model that can generate world simulations based on multiple spatial control inputs of various modalities such as segmentation, depth, and edge. In the design, the spatial conditional scheme is adaptive and customizable. It allows weighting different conditional inputs differently at different spatial locations. This enables highly controllable world generation and finds use in various world-to-world transfer use cases, including Sim2Real. We conduct extensive evaluations to analyze the proposed model and demonstrate its applications for Physical AI, including robotics Sim2Real and autonomous vehicle data enrichment. We further demonstrate an inference scaling strategy to achieve real-time world generation with an NVIDIA GB200 NVL72 rack. To help accelerate research development in the field, we open-source our models and code at https://github.com/nvidia-cosmos/cosmos-transfer1.
Improving the Precision of CNNs for Magnetic Resonance Spectral Modeling
Sep 10, 2024


Abstract:Magnetic resonance spectroscopic imaging is a widely available imaging modality that can non-invasively provide a metabolic profile of the tissue of interest, yet is challenging to integrate clinically. One major reason is the expensive, expert data processing and analysis that is required. Using machine learning to predict MRS-related quantities offers avenues around this problem, but deep learning models bring their own challenges, especially model trust. Current research trends focus primarily on mean error metrics, but comprehensive precision metrics are also needed, e.g. standard deviations, confidence intervals, etc.. This work highlights why more comprehensive error characterization is important and how to improve the precision of CNNs for spectral modeling, a quantitative task. The results highlight advantages and trade-offs of these techniques that should be considered when addressing such regression tasks with CNNs. Detailed insights into the underlying mechanisms of each technique, and how they interact with other techniques, are discussed in depth.
Pay Less On Clinical Images: Asymmetric Multi-Modal Fusion Method For Efficient Multi-Label Skin Lesion Classification
Jul 13, 2024



Abstract:Existing multi-modal approaches primarily focus on enhancing multi-label skin lesion classification performance through advanced fusion modules, often neglecting the associated rise in parameters. In clinical settings, both clinical and dermoscopy images are captured for diagnosis; however, dermoscopy images exhibit more crucial visual features for multi-label skin lesion classification. Motivated by this observation, we introduce a novel asymmetric multi-modal fusion method in this paper for efficient multi-label skin lesion classification. Our fusion method incorporates two innovative schemes. Firstly, we validate the effectiveness of our asymmetric fusion structure. It employs a light and simple network for clinical images and a heavier, more complex one for dermoscopy images, resulting in significant parameter savings compared to the symmetric fusion structure using two identical networks for both modalities. Secondly, in contrast to previous approaches using mutual attention modules for interaction between image modalities, we propose an asymmetric attention module. This module solely leverages clinical image information to enhance dermoscopy image features, considering clinical images as supplementary information in our pipeline. We conduct the extensive experiments on the seven-point checklist dataset. Results demonstrate the generality of our proposed method for both networks and Transformer structures, showcasing its superiority over existing methods We will make our code publicly available.
Single-Shared Network with Prior-Inspired Loss for Parameter-Efficient Multi-Modal Imaging Skin Lesion Classification
Mar 28, 2024

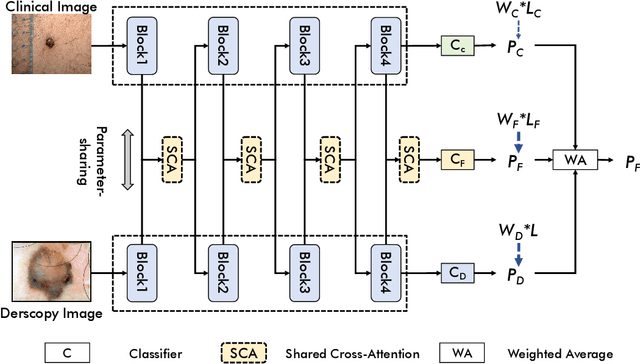

Abstract:In this study, we introduce a multi-modal approach that efficiently integrates multi-scale clinical and dermoscopy features within a single network, thereby substantially reducing model parameters. The proposed method includes three novel fusion schemes. Firstly, unlike current methods that usually employ two individual models for for clinical and dermoscopy modalities, we verified that multimodal feature can be learned by sharing the parameters of encoder while leaving the individual modal-specific classifiers. Secondly, the shared cross-attention module can replace the individual one to efficiently interact between two modalities at multiple layers. Thirdly, different from current methods that equally optimize dermoscopy and clinical branches, inspired by prior knowledge that dermoscopy images play a more significant role than clinical images, we propose a novel biased loss. This loss guides the single-shared network to prioritize dermoscopy information over clinical information, implicitly learning a better joint feature representation for the modal-specific task. Extensive experiments on a well-recognized Seven-Point Checklist (SPC) dataset and a collected dataset demonstrate the effectiveness of our method on both CNN and Transformer structures. Furthermore, our method exhibits superiority in both accuracy and model parameters compared to currently advanced methods.
Sparsity-based background removal for STORM super-resolution images
Jan 15, 2024



Abstract:Single-molecule localization microscopy techniques, like stochastic optical reconstruction microscopy (STORM), visualize biological specimens by stochastically exciting sparse blinking emitters. The raw images suffer from unwanted background fluorescence, which must be removed to achieve super-resolution. We introduce a sparsity-based background removal method by adapting a neural network (SLNet) from a different microscopy domain. The SLNet computes a low-rank representation of the images, and then, by subtracting it from the raw images, the sparse component is computed, representing the frames without the background. We compared our approach with widely used background removal methods, such as the median background removal or the rolling ball algorithm, on two commonly used STORM datasets, one glial cell, and one microtubule dataset. The SLNet delivers STORM frames with less background, leading to higher emitters' localization precision and higher-resolution reconstructed images than commonly used methods. Notably, the SLNet is lightweight and easily trainable (<5 min). Since it is trained in an unsupervised manner, no prior information is required and can be applied to any STORM dataset. We uploaded a pre-trained SLNet to the Bioimage model zoo, easily accessible through ImageJ. Our results show that our sparse decomposition method could be an essential and efficient STORM pre-processing tool.
Joint-Individual Fusion Structure with Fusion Attention Module for Multi-Modal Skin Cancer Classification
Dec 07, 2023



Abstract:Most convolutional neural network (CNN) based methods for skin cancer classification obtain their results using only dermatological images. Although good classification results have been shown, more accurate results can be achieved by considering the patient's metadata, which is valuable clinical information for dermatologists. Current methods only use the simple joint fusion structure (FS) and fusion modules (FMs) for the multi-modal classification methods, there still is room to increase the accuracy by exploring more advanced FS and FM. Therefore, in this paper, we design a new fusion method that combines dermatological images (dermoscopy images or clinical images) and patient metadata for skin cancer classification from the perspectives of FS and FM. First, we propose a joint-individual fusion (JIF) structure that learns the shared features of multi-modality data and preserves specific features simultaneously. Second, we introduce a fusion attention (FA) module that enhances the most relevant image and metadata features based on both the self and mutual attention mechanism to support the decision-making pipeline. We compare the proposed JIF-MMFA method with other state-of-the-art fusion methods on three different public datasets. The results show that our JIF-MMFA method improves the classification results for all tested CNN backbones and performs better than the other fusion methods on the three public datasets, demonstrating our method's effectiveness and robustness
SR-R$^2$KAC: Improving Single Image Defocus Deblurring
Jul 30, 2023



Abstract:We propose an efficient deep learning method for single image defocus deblurring (SIDD) by further exploring inverse kernel properties. Although the current inverse kernel method, i.e., kernel-sharing parallel atrous convolution (KPAC), can address spatially varying defocus blurs, it has difficulty in handling large blurs of this kind. To tackle this issue, we propose a Residual and Recursive Kernel-sharing Atrous Convolution (R$^2$KAC). R$^2$KAC builds on a significant observation of inverse kernels, that is, successive use of inverse-kernel-based deconvolutions with fixed size helps remove unexpected large blurs but produces ringing artifacts. Specifically, on top of kernel-sharing atrous convolutions used to simulate multi-scale inverse kernels, R$^2$KAC applies atrous convolutions recursively to simulate a large inverse kernel. Specifically, on top of kernel-sharing atrous convolutions, R$^2$KAC stacks atrous convolutions recursively to simulate a large inverse kernel. To further alleviate the contingent effect of recursive stacking, i.e., ringing artifacts, we add identity shortcuts between atrous convolutions to simulate residual deconvolutions. Lastly, a scale recurrent module is embedded in the R$^2$KAC network, leading to SR-R$^2$KAC, so that multi-scale information from coarse to fine is exploited to progressively remove the spatially varying defocus blurs. Extensive experimental results show that our method achieves the state-of-the-art performance.
Graph-Ensemble Learning Model for Multi-label Skin Lesion Classification using Dermoscopy and Clinical Images
Jul 04, 2023



Abstract:Many skin lesion analysis (SLA) methods recently focused on developing a multi-modal-based multi-label classification method due to two factors. The first is multi-modal data, i.e., clinical and dermoscopy images, which can provide complementary information to obtain more accurate results than single-modal data. The second one is that multi-label classification, i.e., seven-point checklist (SPC) criteria as an auxiliary classification task can not only boost the diagnostic accuracy of melanoma in the deep learning (DL) pipeline but also provide more useful functions to the clinical doctor as it is commonly used in clinical dermatologist's diagnosis. However, most methods only focus on designing a better module for multi-modal data fusion; few methods explore utilizing the label correlation between SPC and skin disease for performance improvement. This study fills the gap that introduces a Graph Convolution Network (GCN) to exploit prior co-occurrence between each category as a correlation matrix into the DL model for the multi-label classification. However, directly applying GCN degraded the performances in our experiments; we attribute this to the weak generalization ability of GCN in the scenario of insufficient statistical samples of medical data. We tackle this issue by proposing a Graph-Ensemble Learning Model (GELN) that views the prediction from GCN as complementary information of the predictions from the fusion model and adaptively fuses them by a weighted averaging scheme, which can utilize the valuable information from GCN while avoiding its negative influences as much as possible. To evaluate our method, we conduct experiments on public datasets. The results illustrate that our GELN can consistently improve the classification performance on different datasets and that the proposed method can achieve state-of-the-art performance in SPC and diagnosis classification.
Runtime optimization of acquisition trajectories for X-ray computed tomography with a robotic sample holder
Jun 23, 2023Abstract:Tomographic imaging systems are expected to work with a wide range of samples that house complex structures and challenging material compositions, which can influence image quality in a bad way. Complex samples increase total measurement duration and may introduce beam-hardening artifacts that lead to poor reconstruction image quality. This work presents an online trajectory optimization method for an X-ray computed tomography system with a robotic sample holder. The proposed method reduces measurement time and increases reconstruction image quality by generating an optimized spherical trajectory for the given sample without prior knowledge. The trajectory is generated successively at runtime based on intermediate sample measurements. We present experimental results with the robotic sample holder where two sample measurements using an optimized spherical trajectory achieve improved reconstruction quality compared to a conventional spherical trajectory. Our results demonstrate the ability of our system to increase reconstruction image quality and avoid artifacts at runtime when no prior information about the sample is provided.
Fast light-field 3D microscopy with out-of-distribution detection and adaptation through Conditional Normalizing Flows
Jun 14, 2023Abstract:Real-time 3D fluorescence microscopy is crucial for the spatiotemporal analysis of live organisms, such as neural activity monitoring. The eXtended field-of-view light field microscope (XLFM), also known as Fourier light field microscope, is a straightforward, single snapshot solution to achieve this. The XLFM acquires spatial-angular information in a single camera exposure. In a subsequent step, a 3D volume can be algorithmically reconstructed, making it exceptionally well-suited for real-time 3D acquisition and potential analysis. Unfortunately, traditional reconstruction methods (like deconvolution) require lengthy processing times (0.0220 Hz), hampering the speed advantages of the XLFM. Neural network architectures can overcome the speed constraints at the expense of lacking certainty metrics, which renders them untrustworthy for the biomedical realm. This work proposes a novel architecture to perform fast 3D reconstructions of live immobilized zebrafish neural activity based on a conditional normalizing flow. It reconstructs volumes at 8 Hz spanning 512x512x96 voxels, and it can be trained in under two hours due to the small dataset requirements (10 image-volume pairs). Furthermore, normalizing flows allow for exact Likelihood computation, enabling distribution monitoring, followed by out-of-distribution detection and retraining of the system when a novel sample is detected. We evaluate the proposed method on a cross-validation approach involving multiple in-distribution samples (genetically identical zebrafish) and various out-of-distribution ones.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge