Bjoern H Menze
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
Improving the Precision of CNNs for Magnetic Resonance Spectral Modeling
Sep 10, 2024


Abstract:Magnetic resonance spectroscopic imaging is a widely available imaging modality that can non-invasively provide a metabolic profile of the tissue of interest, yet is challenging to integrate clinically. One major reason is the expensive, expert data processing and analysis that is required. Using machine learning to predict MRS-related quantities offers avenues around this problem, but deep learning models bring their own challenges, especially model trust. Current research trends focus primarily on mean error metrics, but comprehensive precision metrics are also needed, e.g. standard deviations, confidence intervals, etc.. This work highlights why more comprehensive error characterization is important and how to improve the precision of CNNs for spectral modeling, a quantitative task. The results highlight advantages and trade-offs of these techniques that should be considered when addressing such regression tasks with CNNs. Detailed insights into the underlying mechanisms of each technique, and how they interact with other techniques, are discussed in depth.
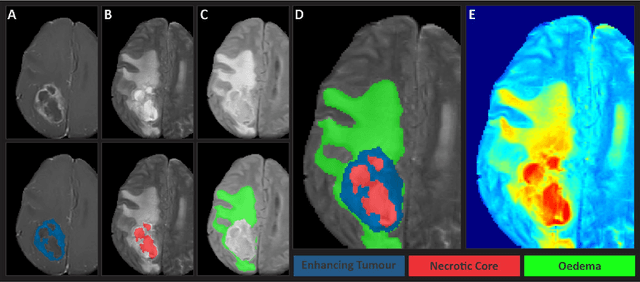
The Brain Tumor Segmentation (BraTS-METS) Challenge 2023: Brain Metastasis Segmentation on Pre-treatment MRI
Jun 01, 2023



Abstract:Clinical monitoring of metastatic disease to the brain can be a laborious and time-consuming process, especially in cases involving multiple metastases when the assessment is performed manually. The Response Assessment in Neuro-Oncology Brain Metastases (RANO-BM) guideline, which utilizes the unidimensional longest diameter, is commonly used in clinical and research settings to evaluate response to therapy in patients with brain metastases. However, accurate volumetric assessment of the lesion and surrounding peri-lesional edema holds significant importance in clinical decision-making and can greatly enhance outcome prediction. The unique challenge in performing segmentations of brain metastases lies in their common occurrence as small lesions. Detection and segmentation of lesions that are smaller than 10 mm in size has not demonstrated high accuracy in prior publications. The brain metastases challenge sets itself apart from previously conducted MICCAI challenges on glioma segmentation due to the significant variability in lesion size. Unlike gliomas, which tend to be larger on presentation scans, brain metastases exhibit a wide range of sizes and tend to include small lesions. We hope that the BraTS-METS dataset and challenge will advance the field of automated brain metastasis detection and segmentation.
The Brain Tumor Segmentation Challenge 2023: Glioma Segmentation in Sub-Saharan Africa Patient Population
May 30, 2023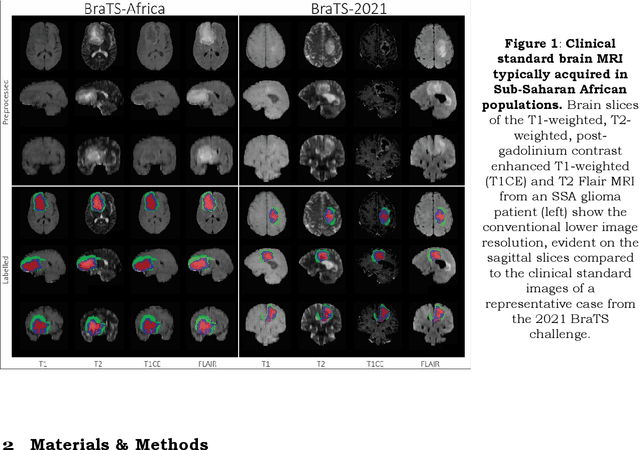
Abstract:Gliomas are the most common type of primary brain tumors. Although gliomas are relatively rare, they are among the deadliest types of cancer, with a survival rate of less than 2 years after diagnosis. Gliomas are challenging to diagnose, hard to treat and inherently resistant to conventional therapy. Years of extensive research to improve diagnosis and treatment of gliomas have decreased mortality rates across the Global North, while chances of survival among individuals in low- and middle-income countries (LMICs) remain unchanged and are significantly worse in Sub-Saharan Africa (SSA) populations. Long-term survival with glioma is associated with the identification of appropriate pathological features on brain MRI and confirmation by histopathology. Since 2012, the Brain Tumor Segmentation (BraTS) Challenge have evaluated state-of-the-art machine learning methods to detect, characterize, and classify gliomas. However, it is unclear if the state-of-the-art methods can be widely implemented in SSA given the extensive use of lower-quality MRI technology, which produces poor image contrast and resolution and more importantly, the propensity for late presentation of disease at advanced stages as well as the unique characteristics of gliomas in SSA (i.e., suspected higher rates of gliomatosis cerebri). Thus, the BraTS-Africa Challenge provides a unique opportunity to include brain MRI glioma cases from SSA in global efforts through the BraTS Challenge to develop and evaluate computer-aided-diagnostic (CAD) methods for the detection and characterization of glioma in resource-limited settings, where the potential for CAD tools to transform healthcare are more likely.
Cross-view Relation Networks for Mammogram Mass Detection
Jul 01, 2019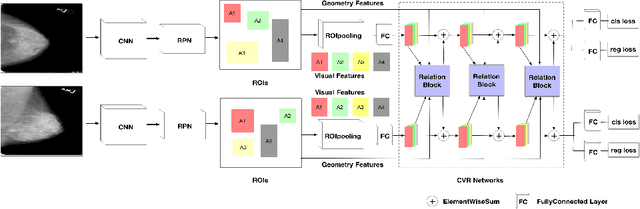
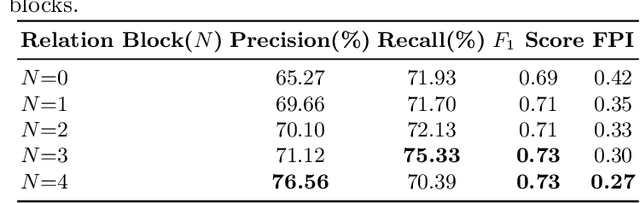
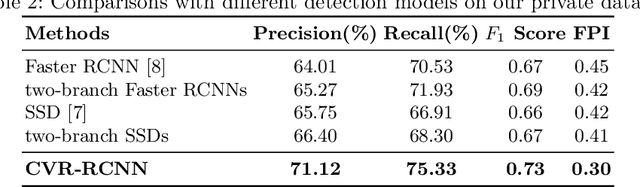
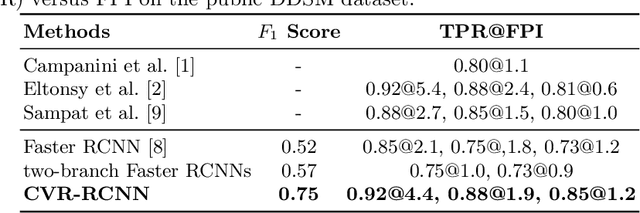
Abstract:Mammogram is the most effective imaging modality for the mass lesion detection of breast cancer at the early stage. The information from the two paired views (i.e., medio-lateral oblique and cranio-caudal) are highly relational and complementary, and this is crucial for doctors' decisions in clinical practice. However, existing mass detection methods do not consider jointly learning effective features from the two relational views. To address this issue, this paper proposes a novel mammogram mass detection framework, termed Cross-View Relation Region-based Convolutional Neural Networks (CVR-RCNN). The proposed CVR-RCNN is expected to capture the latent relation information between the corresponding mass region of interests (ROIs) from the two paired views. Evaluations on a new large-scale private dataset and a public mammogram dataset show that the proposed CVR-RCNN outperforms existing state-of-the-art mass detection methods. Meanwhile, our experimental results suggest that incorporating the relation information across two views helps to train a superior detection model, which is a promising avenue for mammogram mass detection.
Group-Attention Single-Shot Detector (GA-SSD): Finding Pulmonary Nodules in Large-Scale CT Images
Dec 18, 2018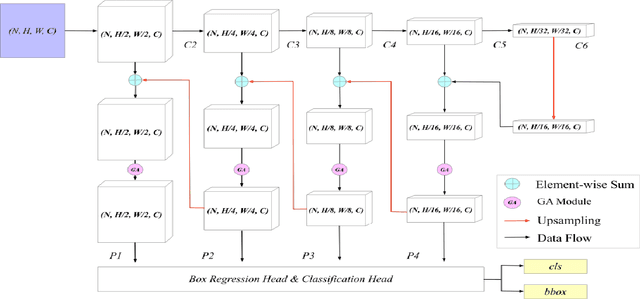
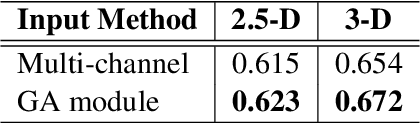
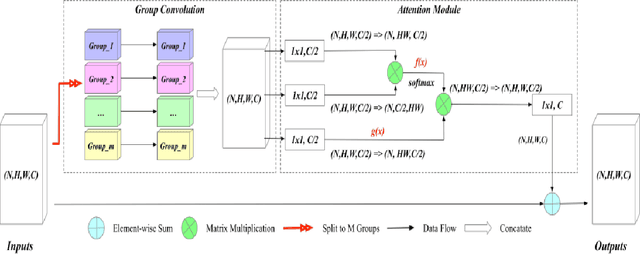

Abstract:Early diagnosis of pulmonary nodules (PNs) can improve the survival rate of patients and yet is a challenging task for radiologists due to the image noise and artifacts in computed tomography (CT) images. In this paper, we propose a novel and effective abnormality detector implementing the attention mechanism and group convolution on 3D single-shot detector (SSD) called group-attention SSD (GA-SSD). We find that group convolution is effective in extracting rich context information between continuous slices, and attention network can learn the target features automatically. We collected a large-scale dataset that contained 4146 CT scans with annotations of varying types and sizes of PNs (even PNs smaller than 3mm were annotated). To the best of our knowledge, this dataset is the largest cohort with relatively complete annotations for PNs detection. Our experimental results show that the proposed group-attention SSD outperforms the classic SSD framework as well as the state-of-the-art 3DCNN, especially on some challenging lesion types.
Qunatification of Metabolites in MR Spectroscopic Imaging using Machine Learning
May 25, 2018



Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is a clinical imaging modality for measuring tissue metabolite levels in-vivo. An accurate estimation of spectral parameters allows for better assessment of spectral quality and metabolite concentration levels. The current gold standard quantification method is the LCModel - a commercial fitting tool. However, this fails for spectra having poor signal-to-noise ratio (SNR) or a large number of artifacts. This paper introduces a framework based on random forest regression for accurate estimation of the output parameters of a model based analysis of MR spectroscopy data. The goal of our proposed framework is to learn the spectral features from a training set comprising of different variations of both simulated and in-vivo brain spectra and then use this learning for the subsequent metabolite quantification. Experiments involve training and testing on simulated and in-vivo human brain spectra. We estimate parameters such as concentration of metabolites and compare our results with that from the LCModel.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge