Jiechao Ma
Correlation-Aware Mutual Learning for Semi-supervised Medical Image Segmentation
Jul 12, 2023Abstract:Semi-supervised learning has become increasingly popular in medical image segmentation due to its ability to leverage large amounts of unlabeled data to extract additional information. However, most existing semi-supervised segmentation methods only focus on extracting information from unlabeled data, disregarding the potential of labeled data to further improve the performance of the model. In this paper, we propose a novel Correlation Aware Mutual Learning (CAML) framework that leverages labeled data to guide the extraction of information from unlabeled data. Our approach is based on a mutual learning strategy that incorporates two modules: the Cross-sample Mutual Attention Module (CMA) and the Omni-Correlation Consistency Module (OCC). The CMA module establishes dense cross-sample correlations among a group of samples, enabling the transfer of label prior knowledge to unlabeled data. The OCC module constructs omni-correlations between the unlabeled and labeled datasets and regularizes dual models by constraining the omni-correlation matrix of each sub-model to be consistent. Experiments on the Atrial Segmentation Challenge dataset demonstrate that our proposed approach outperforms state-of-the-art methods, highlighting the effectiveness of our framework in medical image segmentation tasks. The codes, pre-trained weights, and data are publicly available.
Advancing 3D Medical Image Analysis with Variable Dimension Transform based Supervised 3D Pre-training
Jan 05, 2022



Abstract:The difficulties in both data acquisition and annotation substantially restrict the sample sizes of training datasets for 3D medical imaging applications. As a result, constructing high-performance 3D convolutional neural networks from scratch remains a difficult task in the absence of a sufficient pre-training parameter. Previous efforts on 3D pre-training have frequently relied on self-supervised approaches, which use either predictive or contrastive learning on unlabeled data to build invariant 3D representations. However, because of the unavailability of large-scale supervision information, obtaining semantically invariant and discriminative representations from these learning frameworks remains problematic. In this paper, we revisit an innovative yet simple fully-supervised 3D network pre-training framework to take advantage of semantic supervisions from large-scale 2D natural image datasets. With a redesigned 3D network architecture, reformulated natural images are used to address the problem of data scarcity and develop powerful 3D representations. Comprehensive experiments on four benchmark datasets demonstrate that the proposed pre-trained models can effectively accelerate convergence while also improving accuracy for a variety of 3D medical imaging tasks such as classification, segmentation and detection. In addition, as compared to training from scratch, it can save up to 60% of annotation efforts. On the NIH DeepLesion dataset, it likewise achieves state-of-the-art detection performance, outperforming earlier self-supervised and fully-supervised pre-training approaches, as well as methods that do training from scratch. To facilitate further development of 3D medical models, our code and pre-trained model weights are publicly available at https://github.com/urmagicsmine/CSPR.
Revisiting 3D Context Modeling with Supervised Pre-training for Universal Lesion Detection in CT Slices
Dec 16, 2020



Abstract:Universal lesion detection from computed tomography (CT) slices is important for comprehensive disease screening. Since each lesion can locate in multiple adjacent slices, 3D context modeling is of great significance for developing automated lesion detection algorithms. In this work, we propose a Modified Pseudo-3D Feature Pyramid Network (MP3D FPN) that leverages depthwise separable convolutional filters and a group transform module (GTM) to efficiently extract 3D context enhanced 2D features for universal lesion detection in CT slices. To facilitate faster convergence, a novel 3D network pre-training method is derived using solely large-scale 2D object detection dataset in the natural image domain. We demonstrate that with the novel pre-training method, the proposed MP3D FPN achieves state-of-the-art detection performance on the DeepLesion dataset (3.48% absolute improvement in the sensitivity of FPs@0.5), significantly surpassing the baseline method by up to 6.06% (in MAP@0.5) which adopts 2D convolution for 3D context modeling. Moreover, the proposed 3D pre-trained weights can potentially be used to boost the performance of other 3D medical image analysis tasks.
Invasiveness Prediction of Pulmonary Adenocarcinomas Using Deep Feature Fusion Networks
Sep 21, 2019


Abstract:Early diagnosis of pathological invasiveness of pulmonary adenocarcinomas using computed tomography (CT) imaging would alter the course of treatment of adenocarcinomas and subsequently improve the prognosis. Most of the existing systems use either conventional radiomics features or deep-learning features alone to predict the invasiveness. In this study, we explore the fusion of the two kinds of features and claim that radiomics features can be complementary to deep-learning features. An effective deep feature fusion network is proposed to exploit the complementarity between the two kinds of features, which improves the invasiveness prediction results. We collected a private dataset that contains lung CT scans of 676 patients categorized into four invasiveness types from a collaborating hospital. Evaluations on this dataset demonstrate the effectiveness of our proposal.
Automatic Calcium Scoring in Cardiac and Chest CT Using DenseRAUnet
Jul 26, 2019



Abstract:Cardiovascular disease (CVD) is a common and strong threat to human beings, featuring high prevalence, disability and mortality. The amount of coronary artery calcification (CAC) is an effective factor for CVD risk evaluation. Conventionally, CAC is quantified using ECG-synchronized cardiac CT but rarely from general chest CT scans. However, compared with ECG-synchronized cardiac CT, chest CT is more prevalent and economical in clinical practice. To address this, we propose an automatic method based on Dense U-Net to segment coronary calcium pixels on both types of CT scans. Our contribution is two-fold. First, we propose a novel network called DenseRAUnet, which takes advantage of Dense U-net, ResNet and atrous convolutions. We prove the robustness and generalizability of our model by training it exclusively on chest CT while test on both types of CT scans. Second, we design a loss function combining bootstrap with IoU function to balance foreground and background classes. DenseRAUnet is trained in a 2.5D fashion and tested on a private dataset consisting of 144 scans. Results show an F1-score of 0.75, with 0.83 accuracy of predicting cardiovascular disease risk.
Delving Deep into Liver Focal Lesion Detection: A Preliminary Study
Jul 24, 2019



Abstract:Hepatocellular carcinoma (HCC) is the second most frequent cause of malignancy-related death and is one of the diseases with the highest incidence in the world. Because the liver is the only organ in the human body that is supplied by two major vessels: the hepatic artery and the portal vein, various types of malignant tumors can spread from other organs to the liver. And due to the liver masses' heterogeneous and diffusive shape, the tumor lesions are very difficult to be recognized, thus automatic lesion detection is necessary for the doctors with huge workloads. To assist doctors, this work uses the existing large-scale annotation medical image data to delve deep into liver lesion detection from multiple directions. To solve technical difficulties, such as the image-recognition task, traditional deep learning with convolution neural networks (CNNs) has been widely applied in recent years. However, this kind of neural network, such as Faster Regions with CNN features (R-CNN), cannot leverage the spatial information because it is applied in natural images (2D) rather than medical images (3D), such as computed tomography (CT) images. To address this issue, we propose a novel algorithm that is appropriate for liver CT imaging. Furthermore, according to radiologists' experience in clinical diagnosis and the characteristics of CT images of liver cancer, a liver cancer-detection framework with CNN, including image processing, feature extraction, region proposal, image registration, and classification recognition, was proposed to facilitate the effective detection of liver lesions.
Cross-view Relation Networks for Mammogram Mass Detection
Jul 01, 2019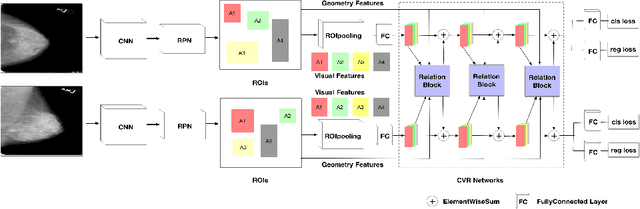
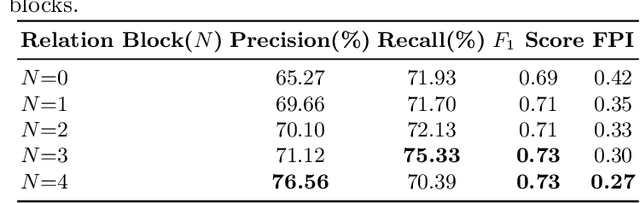
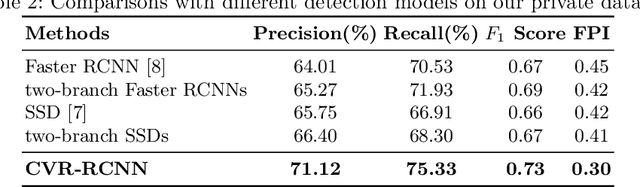
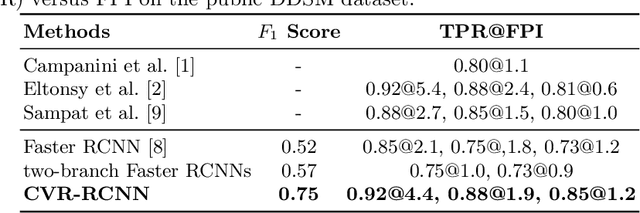
Abstract:Mammogram is the most effective imaging modality for the mass lesion detection of breast cancer at the early stage. The information from the two paired views (i.e., medio-lateral oblique and cranio-caudal) are highly relational and complementary, and this is crucial for doctors' decisions in clinical practice. However, existing mass detection methods do not consider jointly learning effective features from the two relational views. To address this issue, this paper proposes a novel mammogram mass detection framework, termed Cross-View Relation Region-based Convolutional Neural Networks (CVR-RCNN). The proposed CVR-RCNN is expected to capture the latent relation information between the corresponding mass region of interests (ROIs) from the two paired views. Evaluations on a new large-scale private dataset and a public mammogram dataset show that the proposed CVR-RCNN outperforms existing state-of-the-art mass detection methods. Meanwhile, our experimental results suggest that incorporating the relation information across two views helps to train a superior detection model, which is a promising avenue for mammogram mass detection.
Towards Photo-Realistic Visible Watermark Removal with Conditional Generative Adversarial Networks
May 31, 2019



Abstract:Visible watermark plays an important role in image copyright protection and the robustness of a visible watermark to an attack is shown to be essential. To evaluate and improve the effectiveness of watermark, watermark removal attracts increasing attention and becomes a hot research top. Current methods cast the watermark removal as an image-to-image translation problem where the encode-decode architectures with pixel-wise loss are adopted to transfer the transparent watermarked pixels into unmarked pixels. However, when a number of realistic images are presented, the watermarks are more likely to be unknown and diverse (i.e., the watermarks might be opaque or semi-transparent; the category and pattern of watermarks are unknown). When applying existing methods to the real-world scenarios, they mostly can not satisfactorily reconstruct the hidden information obscured under the complex and various watermarks (i.e., the residual watermark traces remain and the reconstructed images lack reality). To address this difficulty, in this paper, we present a new watermark processing framework using the conditional generative adversarial networks (cGANs) for visible watermark removal in the real-world application. The proposed method enables the watermark removal solution to be more closed to the photo-realistic reconstruction using a patch-based discriminator conditioned on the watermarked images, which is adversarially trained to differentiate the difference between the recovered images and original watermark-free images. Extensive experimental results on a large-scale visible watermark dataset demonstrate the effectiveness of the proposed method and clearly indicate that our proposed approach can produce more photo-realistic and convincing results compared with the state-of-the-art methods.
Group-Attention Single-Shot Detector (GA-SSD): Finding Pulmonary Nodules in Large-Scale CT Images
Dec 18, 2018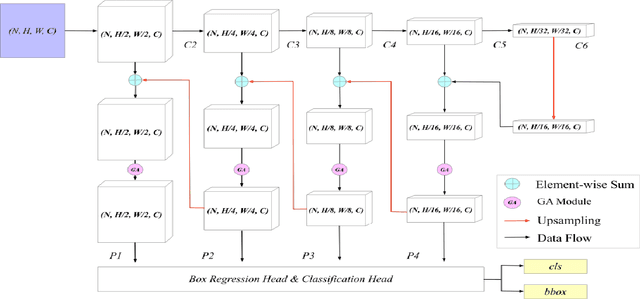
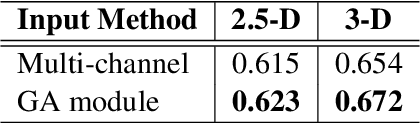
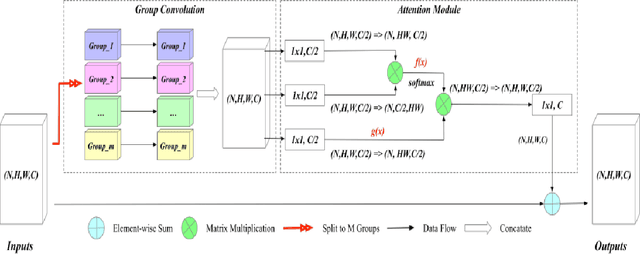

Abstract:Early diagnosis of pulmonary nodules (PNs) can improve the survival rate of patients and yet is a challenging task for radiologists due to the image noise and artifacts in computed tomography (CT) images. In this paper, we propose a novel and effective abnormality detector implementing the attention mechanism and group convolution on 3D single-shot detector (SSD) called group-attention SSD (GA-SSD). We find that group convolution is effective in extracting rich context information between continuous slices, and attention network can learn the target features automatically. We collected a large-scale dataset that contained 4146 CT scans with annotations of varying types and sizes of PNs (even PNs smaller than 3mm were annotated). To the best of our knowledge, this dataset is the largest cohort with relatively complete annotations for PNs detection. Our experimental results show that the proposed group-attention SSD outperforms the classic SSD framework as well as the state-of-the-art 3DCNN, especially on some challenging lesion types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge