Viren Jain
Discovering Mechanistic Models of Neural Activity: System Identification in an in Silico Zebrafish
Feb 04, 2026Abstract:Constructing mechanistic models of neural circuits is a fundamental goal of neuroscience, yet verifying such models is limited by the lack of ground truth. To rigorously test model discovery, we establish an in silico testbed using neuromechanical simulations of a larval zebrafish as a transparent ground truth. We find that LLM-based tree search autonomously discovers predictive models that significantly outperform established forecasting baselines. Conditioning on sensory drive is necessary but not sufficient for faithful system identification, as models exploit statistical shortcuts. Structural priors prove essential for enabling robust out-of-distribution generalization and recovery of interpretable mechanistic models. Our insights provide guidance for modeling real-world neural recordings and offer a broader template for AI-driven scientific discovery.
ZAPBench: A Benchmark for Whole-Brain Activity Prediction in Zebrafish
Mar 04, 2025



Abstract:Data-driven benchmarks have led to significant progress in key scientific modeling domains including weather and structural biology. Here, we introduce the Zebrafish Activity Prediction Benchmark (ZAPBench) to measure progress on the problem of predicting cellular-resolution neural activity throughout an entire vertebrate brain. The benchmark is based on a novel dataset containing 4d light-sheet microscopy recordings of over 70,000 neurons in a larval zebrafish brain, along with motion stabilized and voxel-level cell segmentations of these data that facilitate development of a variety of forecasting methods. Initial results from a selection of time series and volumetric video modeling approaches achieve better performance than naive baseline methods, but also show room for further improvement. The specific brain used in the activity recording is also undergoing synaptic-level anatomical mapping, which will enable future integration of detailed structural information into forecasting methods.
How to Build the Virtual Cell with Artificial Intelligence: Priorities and Opportunities
Sep 18, 2024

Abstract:The cell is arguably the smallest unit of life and is central to understanding biology. Accurate modeling of cells is important for this understanding as well as for determining the root causes of disease. Recent advances in artificial intelligence (AI), combined with the ability to generate large-scale experimental data, present novel opportunities to model cells. Here we propose a vision of AI-powered Virtual Cells, where robust representations of cells and cellular systems under different conditions are directly learned from growing biological data across measurements and scales. We discuss desired capabilities of AI Virtual Cells, including generating universal representations of biological entities across scales, and facilitating interpretable in silico experiments to predict and understand their behavior using Virtual Instruments. We further address the challenges, opportunities and requirements to realize this vision including data needs, evaluation strategies, and community standards and engagement to ensure biological accuracy and broad utility. We envision a future where AI Virtual Cells help identify new drug targets, predict cellular responses to perturbations, as well as scale hypothesis exploration. With open science collaborations across the biomedical ecosystem that includes academia, philanthropy, and the biopharma and AI industries, a comprehensive predictive understanding of cell mechanisms and interactions is within reach.
Adversarial Image Alignment and Interpolation
Jun 30, 2017

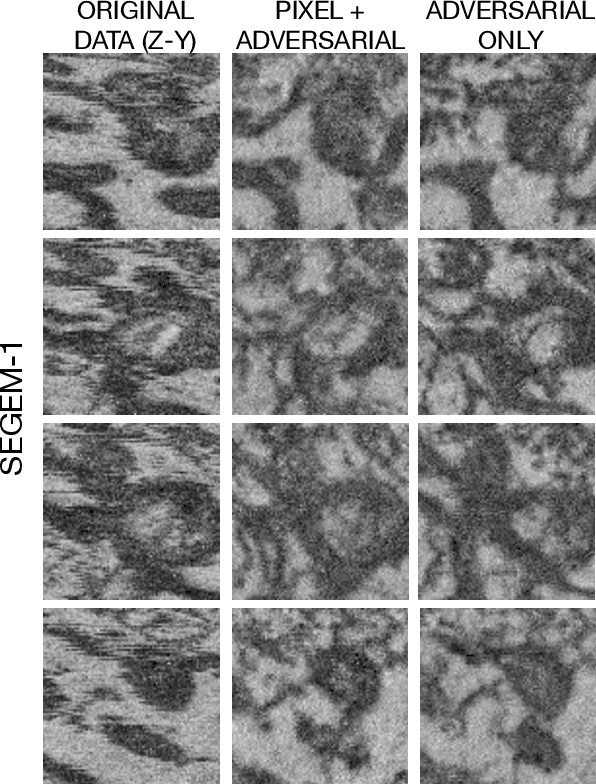

Abstract:Volumetric (3d) images are acquired for many scientific and biomedical purposes using imaging methods such as serial section microscopy, CT scans, and MRI. A frequent step in the analysis and reconstruction of such data is the alignment and registration of images that were acquired in succession along a spatial or temporal dimension. For example, in serial section electron microscopy, individual 2d sections are imaged via electron microscopy and then must be aligned to one another in order to produce a coherent 3d volume. State of the art approaches find image correspondences derived from patch matching and invariant feature detectors, and then solve optimization problems that rigidly or elastically deform series of images into an aligned volume. Here we show how fully convolutional neural networks trained with an adversarial loss function can be used for two tasks: (1) synthesis of missing or damaged image data from adjacent sections, and (2) fine-scale alignment of block-face electron microscopy data. Finally, we show how these two capabilities can be combined in order to produce artificial isotropic volumes from anisotropic image volumes using a super-resolution adversarial alignment and interpolation approach.
Superhuman Accuracy on the SNEMI3D Connectomics Challenge
May 31, 2017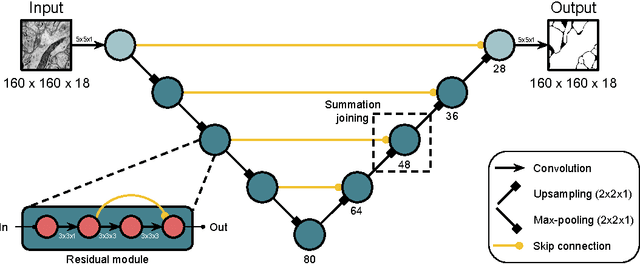
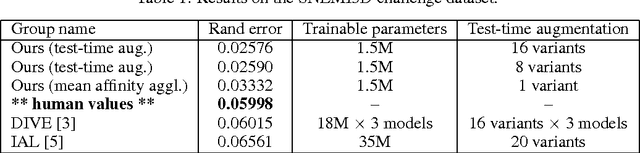
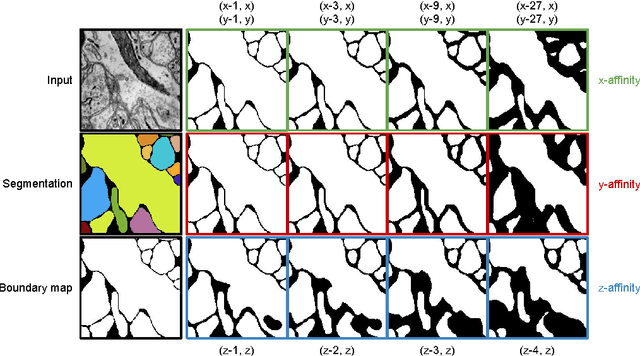
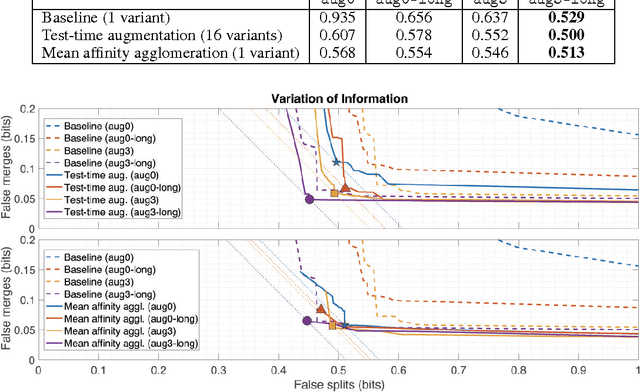
Abstract:For the past decade, convolutional networks have been used for 3D reconstruction of neurons from electron microscopic (EM) brain images. Recent years have seen great improvements in accuracy, as evidenced by submissions to the SNEMI3D benchmark challenge. Here we report the first submission to surpass the estimate of human accuracy provided by the SNEMI3D leaderboard. A variant of 3D U-Net is trained on a primary task of predicting affinities between nearest neighbor voxels, and an auxiliary task of predicting long-range affinities. The training data is augmented by simulated image defects. The nearest neighbor affinities are used to create an oversegmentation, and then supervoxels are greedily agglomerated based on mean affinity. The resulting SNEMI3D score exceeds the estimate of human accuracy by a large margin. While one should be cautious about extrapolating from the SNEMI3D benchmark to real-world accuracy of large-scale neural circuit reconstruction, our result inspires optimism that the goal of full automation may be realizable in the future.
Morphological Error Detection in 3D Segmentations
May 30, 2017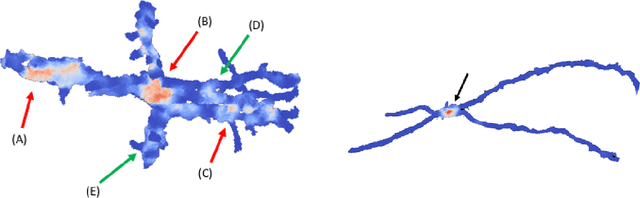
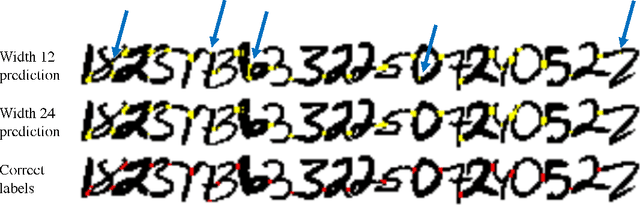
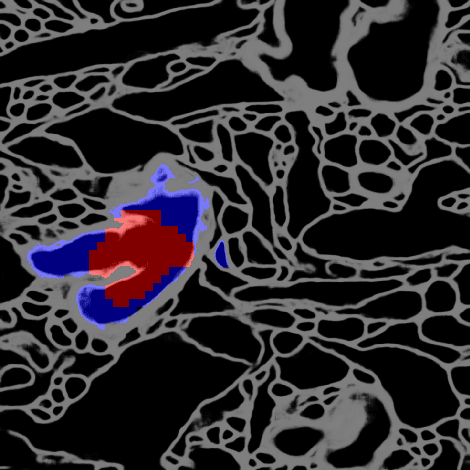
Abstract:Deep learning algorithms for connectomics rely upon localized classification, rather than overall morphology. This leads to a high incidence of erroneously merged objects. Humans, by contrast, can easily detect such errors by acquiring intuition for the correct morphology of objects. Biological neurons have complicated and variable shapes, which are challenging to learn, and merge errors take a multitude of different forms. We present an algorithm, MergeNet, that shows 3D ConvNets can, in fact, detect merge errors from high-level neuronal morphology. MergeNet follows unsupervised training and operates across datasets. We demonstrate the performance of MergeNet both on a variety of connectomics data and on a dataset created from merged MNIST images.
Flood-Filling Networks
Nov 01, 2016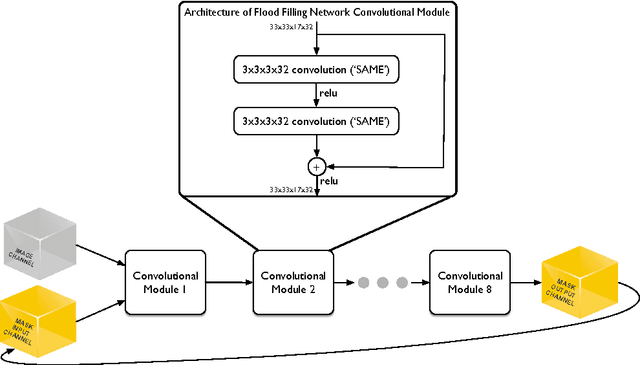
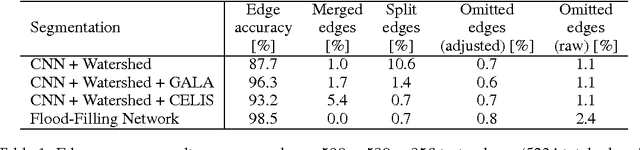
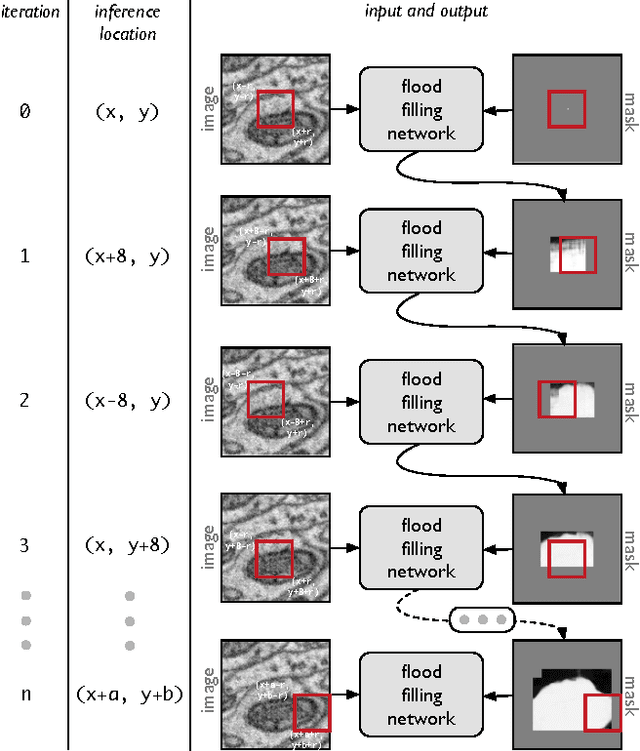
Abstract:State-of-the-art image segmentation algorithms generally consist of at least two successive and distinct computations: a boundary detection process that uses local image information to classify image locations as boundaries between objects, followed by a pixel grouping step such as watershed or connected components that clusters pixels into segments. Prior work has varied the complexity and approach employed in these two steps, including the incorporation of multi-layer neural networks to perform boundary prediction, and the use of global optimizations during pixel clustering. We propose a unified and end-to-end trainable machine learning approach, flood-filling networks, in which a recurrent 3d convolutional network directly produces individual segments from a raw image. The proposed approach robustly segments images with an unknown and variable number of objects as well as highly variable object sizes. We demonstrate the approach on a challenging 3d image segmentation task, connectomic reconstruction from volume electron microscopy data, on which flood-filling neural networks substantially improve accuracy over other state-of-the-art methods. The proposed approach can replace complex multi-step segmentation pipelines with a single neural network that is learned end-to-end.
Combinatorial Energy Learning for Image Segmentation
Sep 23, 2016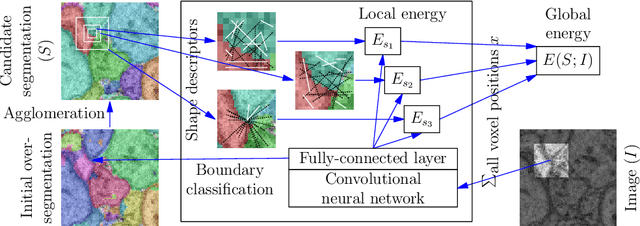
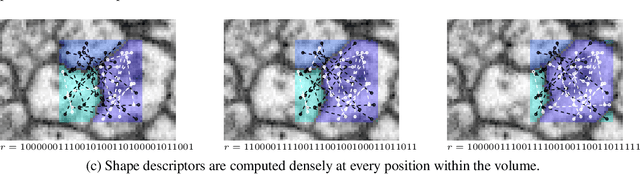
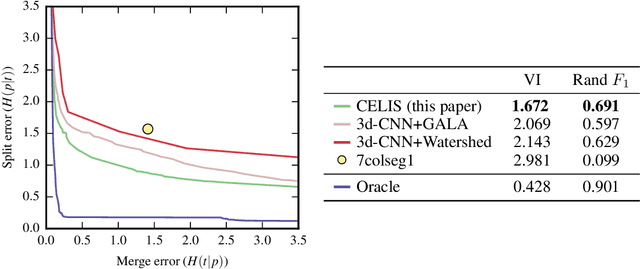
Abstract:We introduce a new machine learning approach for image segmentation that uses a neural network to model the conditional energy of a segmentation given an image. Our approach, combinatorial energy learning for image segmentation (CELIS) places a particular emphasis on modeling the inherent combinatorial nature of dense image segmentation problems. We propose efficient algorithms for learning deep neural networks to model the energy function, and for local optimization of this energy in the space of supervoxel agglomerations. We extensively evaluate our method on a publicly available 3-D microscopy dataset with 25 billion voxels of ground truth data. On an 11 billion voxel test set, we find that our method improves volumetric reconstruction accuracy by more than 20% as compared to two state-of-the-art baseline methods: graph-based segmentation of the output of a 3-D convolutional neural network trained to predict boundaries, as well as a random forest classifier trained to agglomerate supervoxels that were generated by a 3-D convolutional neural network.
Learned versus Hand-Designed Feature Representations for 3d Agglomeration
Dec 20, 2013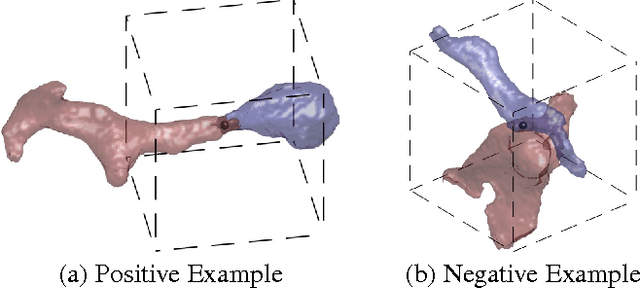
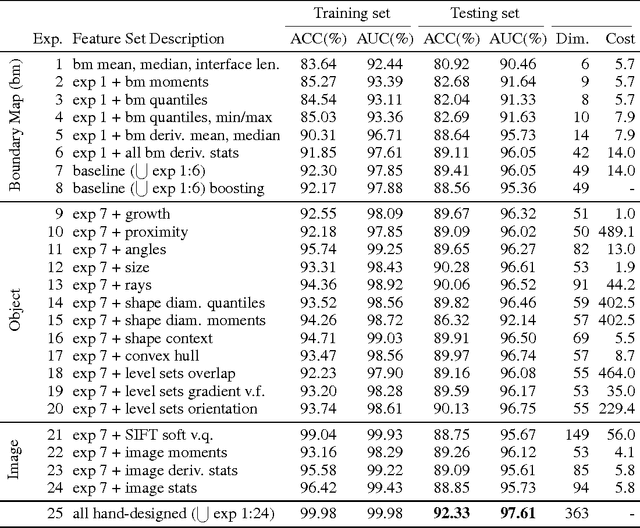
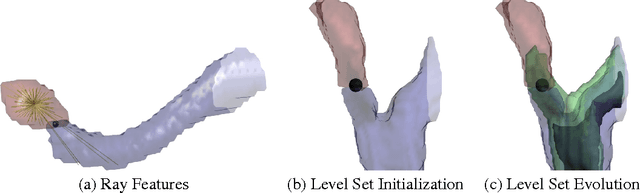
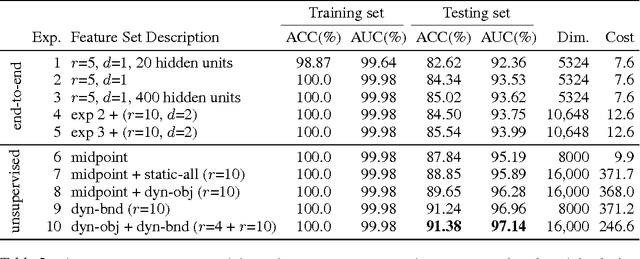
Abstract:For image recognition and labeling tasks, recent results suggest that machine learning methods that rely on manually specified feature representations may be outperformed by methods that automatically derive feature representations based on the data. Yet for problems that involve analysis of 3d objects, such as mesh segmentation, shape retrieval, or neuron fragment agglomeration, there remains a strong reliance on hand-designed feature descriptors. In this paper, we evaluate a large set of hand-designed 3d feature descriptors alongside features learned from the raw data using both end-to-end and unsupervised learning techniques, in the context of agglomeration of 3d neuron fragments. By combining unsupervised learning techniques with a novel dynamic pooling scheme, we show how pure learning-based methods are for the first time competitive with hand-designed 3d shape descriptors. We investigate data augmentation strategies for dramatically increasing the size of the training set, and show how combining both learned and hand-designed features leads to the highest accuracy.
Deep and Wide Multiscale Recursive Networks for Robust Image Labeling
Dec 06, 2013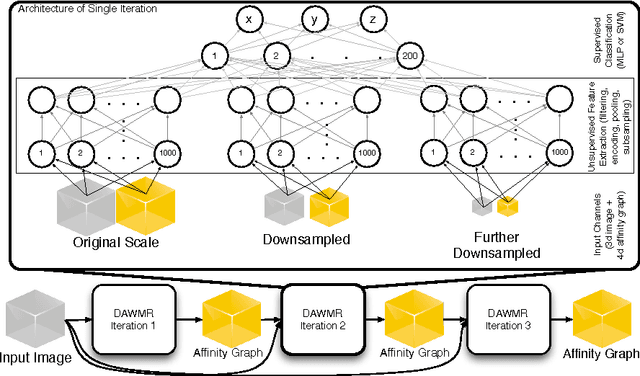
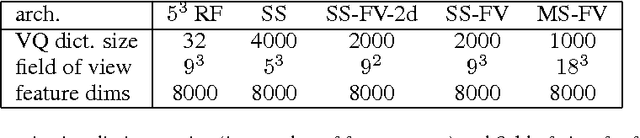
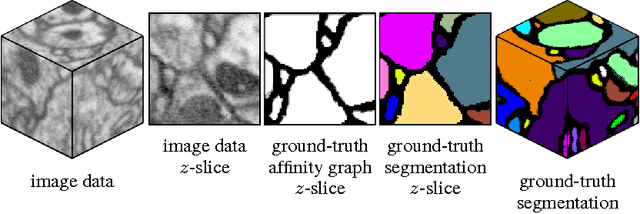
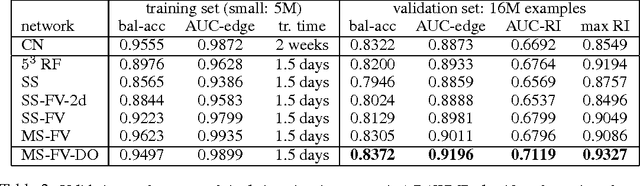
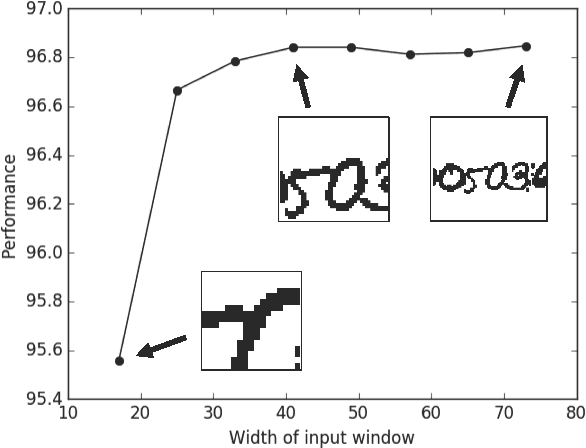
Abstract:Feedforward multilayer networks trained by supervised learning have recently demonstrated state of the art performance on image labeling problems such as boundary prediction and scene parsing. As even very low error rates can limit practical usage of such systems, methods that perform closer to human accuracy remain desirable. In this work, we propose a new type of network with the following properties that address what we hypothesize to be limiting aspects of existing methods: (1) a `wide' structure with thousands of features, (2) a large field of view, (3) recursive iterations that exploit statistical dependencies in label space, and (4) a parallelizable architecture that can be trained in a fraction of the time compared to benchmark multilayer convolutional networks. For the specific image labeling problem of boundary prediction, we also introduce a novel example weighting algorithm that improves segmentation accuracy. Experiments in the challenging domain of connectomic reconstruction of neural circuity from 3d electron microscopy data show that these "Deep And Wide Multiscale Recursive" (DAWMR) networks lead to new levels of image labeling performance. The highest performing architecture has twelve layers, interwoven supervised and unsupervised stages, and uses an input field of view of 157,464 voxels ($54^3$) to make a prediction at each image location. We present an associated open source software package that enables the simple and flexible creation of DAWMR networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge