Gary B. Huang
Fully-Automatic Synapse Prediction and Validation on a Large Data Set
Apr 11, 2016



Abstract:Extracting a connectome from an electron microscopy (EM) data set requires identification of neurons and determination of synapses between neurons. As manual extraction of this information is very time-consuming, there has been extensive research effort to automatically segment the neurons to help guide and eventually replace manual tracing. Until recently, there has been comparatively less research on automatically detecting the actual synapses between neurons. This discrepancy can, in part, be attributed to several factors: obtaining neuronal shapes is a prerequisite first step in extracting a connectome, manual tracing is much more time-consuming than annotating synapses, and neuronal contact area can be used as a proxy for synapses in determining connections. However, recent research has demonstrated that contact area alone is not a sufficient predictor of synaptic connection. Moreover, as segmentation has improved, we have observed that synapse annotation is consuming a more significant fraction of overall reconstruction time. This ratio will only get worse as segmentation improves, gating overall possible speed-up. Therefore, we address this problem by developing algorithms that automatically detect pre-synaptic neurons and their post-synaptic partners. In particular, pre-synaptic structures are detected using a Deep and Wide Multiscale Recursive Network, and post-synaptic partners are detected using a MLP with features conditioned on the local segmentation. This work is novel because it requires minimal amount of training, leverages advances in image segmentation directly, and provides a complete solution for polyadic synapse detection. We further introduce novel metrics to evaluate our algorithm on connectomes of meaningful size. These metrics demonstrate that complete automatic prediction can be used to effectively characterize most connectivity correctly.
Annotating Synapses in Large EM Datasets
Dec 04, 2014



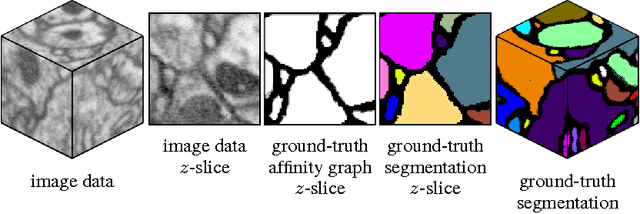
Abstract:Reconstructing neuronal circuits at the level of synapses is a central problem in neuroscience and becoming a focus of the emerging field of connectomics. To date, electron microscopy (EM) is the most proven technique for identifying and quantifying synaptic connections. As advances in EM make acquiring larger datasets possible, subsequent manual synapse identification ({\em i.e.}, proofreading) for deciphering a connectome becomes a major time bottleneck. Here we introduce a large-scale, high-throughput, and semi-automated methodology to efficiently identify synapses. We successfully applied our methodology to the Drosophila medulla optic lobe, annotating many more synapses than previous connectome efforts. Our approaches are extensible and will make the often complicated process of synapse identification accessible to a wider-community of potential proofreaders.
Identifying Synapses Using Deep and Wide Multiscale Recursive Networks
Sep 05, 2014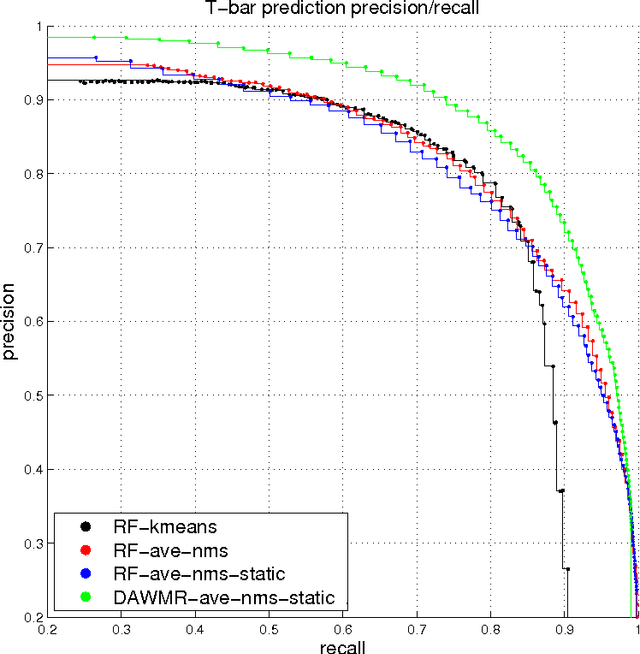
Abstract:In this work, we propose a learning framework for identifying synapses using a deep and wide multi-scale recursive (DAWMR) network, previously considered in image segmentation applications. We apply this approach on electron microscopy data from invertebrate fly brain tissue. By learning features directly from the data, we are able to achieve considerable improvements over existing techniques that rely on a small set of hand-designed features. We show that this system can reduce the amount of manual annotation required, in both acquisition of training data as well as verification of inferred detections.
Learned versus Hand-Designed Feature Representations for 3d Agglomeration
Dec 20, 2013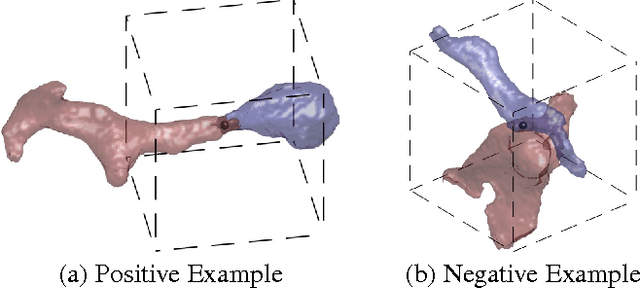
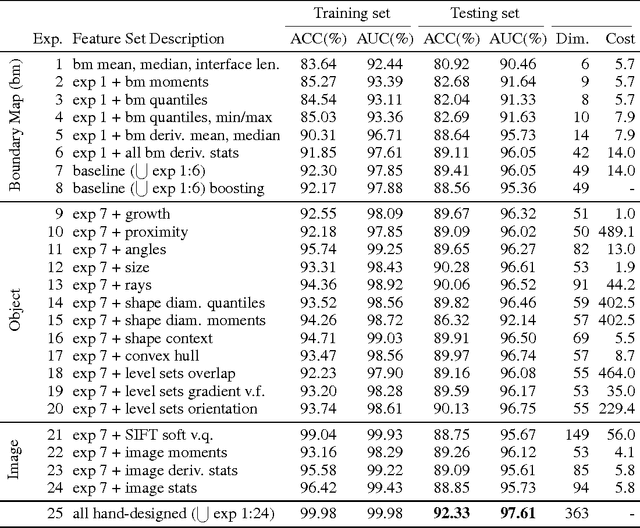
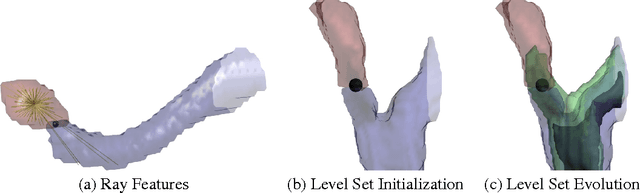
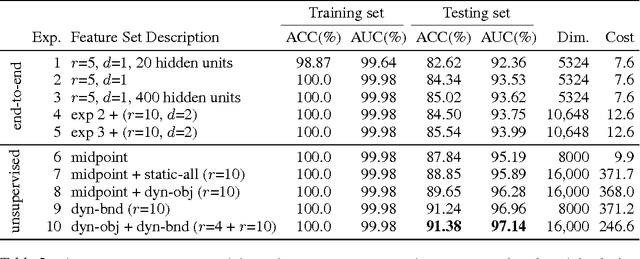
Abstract:For image recognition and labeling tasks, recent results suggest that machine learning methods that rely on manually specified feature representations may be outperformed by methods that automatically derive feature representations based on the data. Yet for problems that involve analysis of 3d objects, such as mesh segmentation, shape retrieval, or neuron fragment agglomeration, there remains a strong reliance on hand-designed feature descriptors. In this paper, we evaluate a large set of hand-designed 3d feature descriptors alongside features learned from the raw data using both end-to-end and unsupervised learning techniques, in the context of agglomeration of 3d neuron fragments. By combining unsupervised learning techniques with a novel dynamic pooling scheme, we show how pure learning-based methods are for the first time competitive with hand-designed 3d shape descriptors. We investigate data augmentation strategies for dramatically increasing the size of the training set, and show how combining both learned and hand-designed features leads to the highest accuracy.
Deep and Wide Multiscale Recursive Networks for Robust Image Labeling
Dec 06, 2013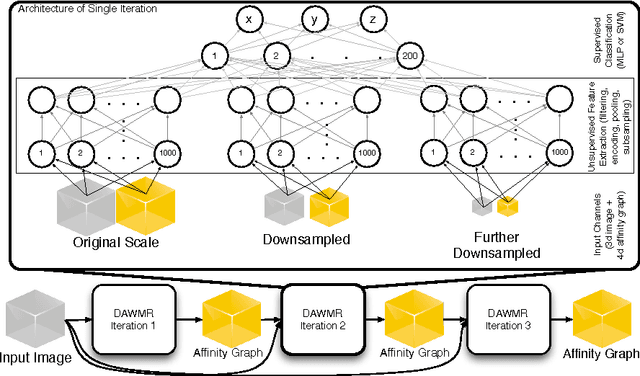
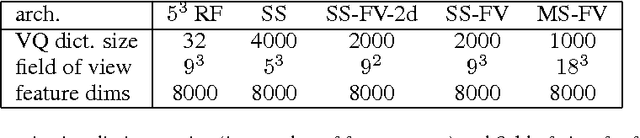
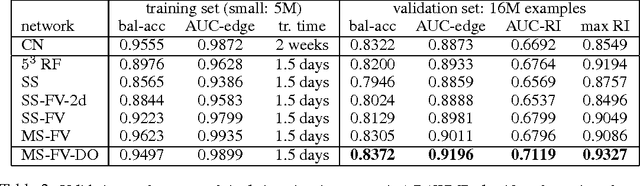
Abstract:Feedforward multilayer networks trained by supervised learning have recently demonstrated state of the art performance on image labeling problems such as boundary prediction and scene parsing. As even very low error rates can limit practical usage of such systems, methods that perform closer to human accuracy remain desirable. In this work, we propose a new type of network with the following properties that address what we hypothesize to be limiting aspects of existing methods: (1) a `wide' structure with thousands of features, (2) a large field of view, (3) recursive iterations that exploit statistical dependencies in label space, and (4) a parallelizable architecture that can be trained in a fraction of the time compared to benchmark multilayer convolutional networks. For the specific image labeling problem of boundary prediction, we also introduce a novel example weighting algorithm that improves segmentation accuracy. Experiments in the challenging domain of connectomic reconstruction of neural circuity from 3d electron microscopy data show that these "Deep And Wide Multiscale Recursive" (DAWMR) networks lead to new levels of image labeling performance. The highest performing architecture has twelve layers, interwoven supervised and unsupervised stages, and uses an input field of view of 157,464 voxels ($54^3$) to make a prediction at each image location. We present an associated open source software package that enables the simple and flexible creation of DAWMR networks.
Bounding the Probability of Error for High Precision Recognition
Jul 02, 2009
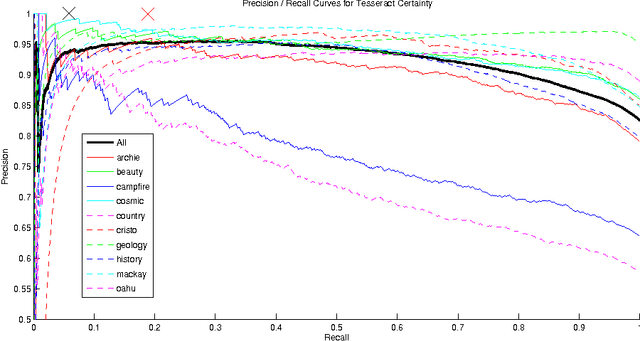
Abstract:We consider models for which it is important, early in processing, to estimate some variables with high precision, but perhaps at relatively low rates of recall. If some variables can be identified with near certainty, then they can be conditioned upon, allowing further inference to be done efficiently. Specifically, we consider optical character recognition (OCR) systems that can be bootstrapped by identifying a subset of correctly translated document words with very high precision. This "clean set" is subsequently used as document-specific training data. While many current OCR systems produce measures of confidence for the identity of each letter or word, thresholding these confidence values, even at very high values, still produces some errors. We introduce a novel technique for identifying a set of correct words with very high precision. Rather than estimating posterior probabilities, we bound the probability that any given word is incorrect under very general assumptions, using an approximate worst case analysis. As a result, the parameters of the model are nearly irrelevant, and we are able to identify a subset of words, even in noisy documents, of which we are highly confident. On our set of 10 documents, we are able to identify about 6% of the words on average without making a single error. This ability to produce word lists with very high precision allows us to use a family of models which depends upon such clean word lists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge