Jonathan Zung
An Error Detection and Correction Framework for Connectomics
Dec 03, 2017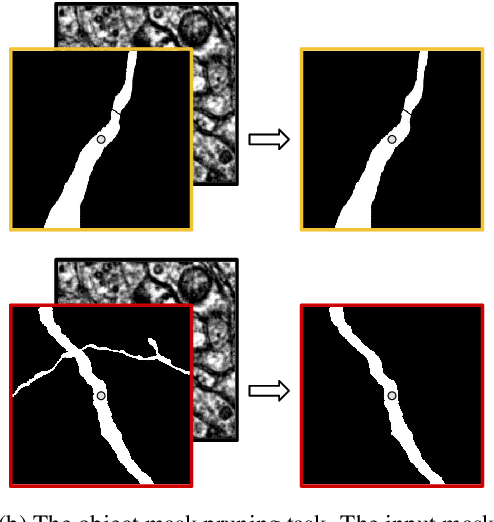
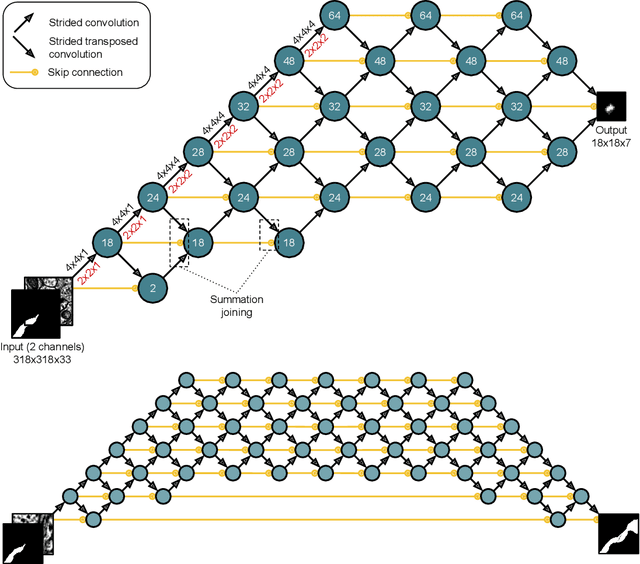

Abstract:We define and study error detection and correction tasks that are useful for 3D reconstruction of neurons from electron microscopic imagery, and for image segmentation more generally. Both tasks take as input the raw image and a binary mask representing a candidate object. For the error detection task, the desired output is a map of split and merge errors in the object. For the error correction task, the desired output is the true object. We call this object mask pruning, because the candidate object mask is assumed to be a superset of the true object. We train multiscale 3D convolutional networks to perform both tasks. We find that the error-detecting net can achieve high accuracy. The accuracy of the error-correcting net is enhanced if its input object mask is "advice" (union of erroneous objects) from the error-detecting net.
Superhuman Accuracy on the SNEMI3D Connectomics Challenge
May 31, 2017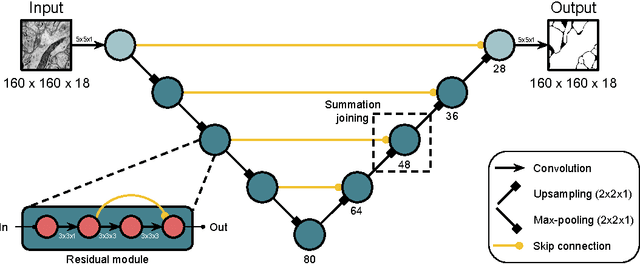
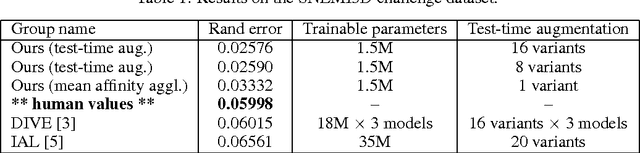
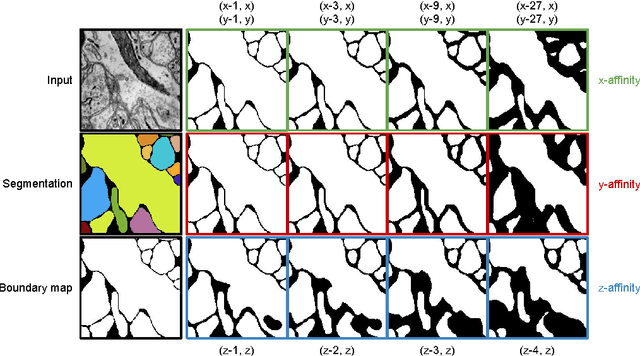
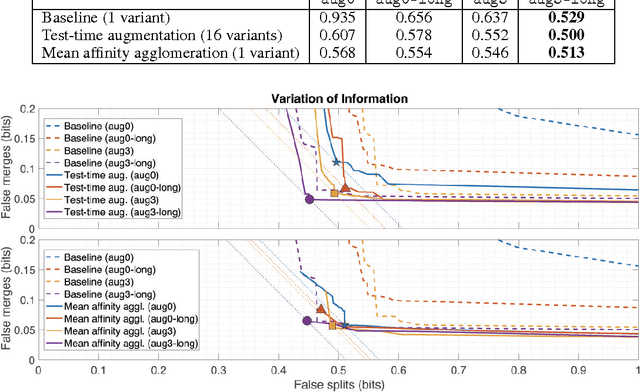
Abstract:For the past decade, convolutional networks have been used for 3D reconstruction of neurons from electron microscopic (EM) brain images. Recent years have seen great improvements in accuracy, as evidenced by submissions to the SNEMI3D benchmark challenge. Here we report the first submission to surpass the estimate of human accuracy provided by the SNEMI3D leaderboard. A variant of 3D U-Net is trained on a primary task of predicting affinities between nearest neighbor voxels, and an auxiliary task of predicting long-range affinities. The training data is augmented by simulated image defects. The nearest neighbor affinities are used to create an oversegmentation, and then supervoxels are greedily agglomerated based on mean affinity. The resulting SNEMI3D score exceeds the estimate of human accuracy by a large margin. While one should be cautious about extrapolating from the SNEMI3D benchmark to real-world accuracy of large-scale neural circuit reconstruction, our result inspires optimism that the goal of full automation may be realizable in the future.
Deep Learning Improves Template Matching by Normalized Cross Correlation
May 24, 2017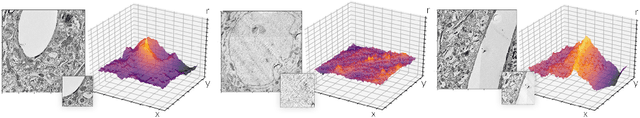

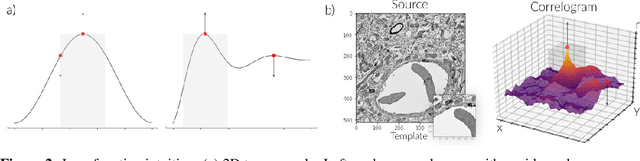
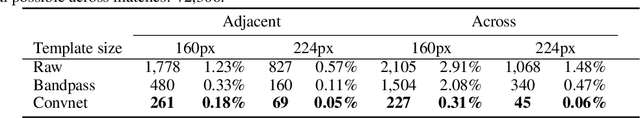
Abstract:Template matching by normalized cross correlation (NCC) is widely used for finding image correspondences. We improve the robustness of this algorithm by preprocessing images with "siamese" convolutional networks trained to maximize the contrast between NCC values of true and false matches. The improvement is quantified using patches of brain images from serial section electron microscopy. Relative to a parameter-tuned bandpass filter, siamese convolutional networks significantly reduce false matches. Furthermore, all false matches can be eliminated by removing a tiny fraction of all matches based on NCC values. The improved accuracy of our method could be essential for connectomics, because emerging petascale datasets may require billions of template matches to assemble 2D images of serial sections into a 3D image stack. Our method is also expected to generalize to many other computer vision applications that use NCC template matching to find image correspondences.
A correlation game for unsupervised learning yields computational interpretations of Hebbian excitation, anti-Hebbian inhibition, and synapse elimination
Apr 03, 2017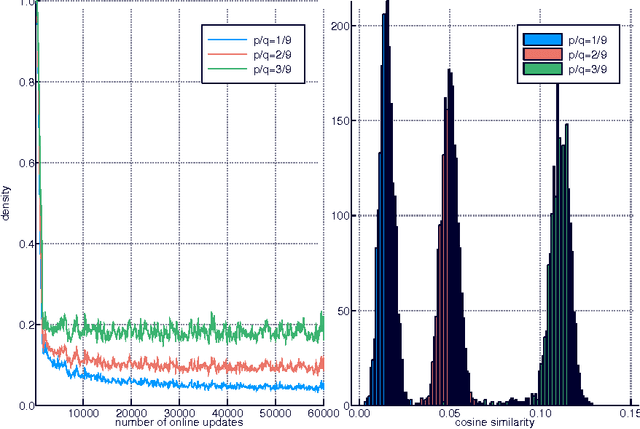
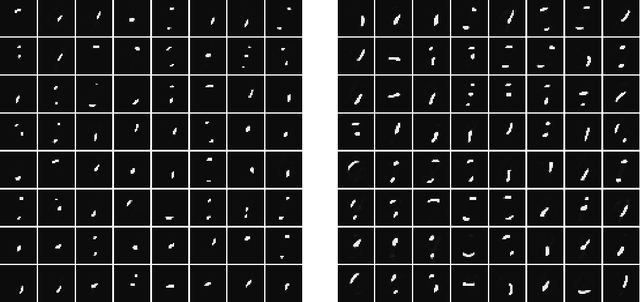
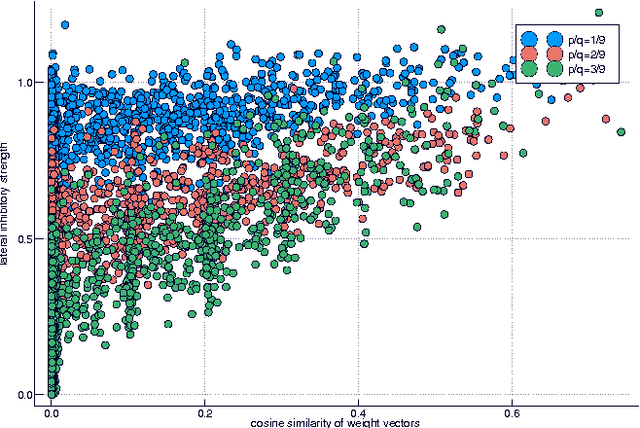
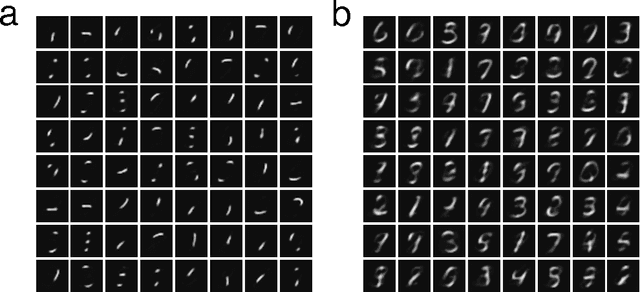
Abstract:Much has been learned about plasticity of biological synapses from empirical studies. Hebbian plasticity is driven by correlated activity of presynaptic and postsynaptic neurons. Synapses that converge onto the same neuron often behave as if they compete for a fixed resource; some survive the competition while others are eliminated. To provide computational interpretations of these aspects of synaptic plasticity, we formulate unsupervised learning as a zero-sum game between Hebbian excitation and anti-Hebbian inhibition in a neural network model. The game formalizes the intuition that Hebbian excitation tries to maximize correlations of neurons with their inputs, while anti-Hebbian inhibition tries to decorrelate neurons from each other. We further include a model of synaptic competition, which enables a neuron to eliminate all connections except those from its most strongly correlated inputs. Through empirical studies, we show that this facilitates the learning of sensory features that resemble parts of objects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge