Davit Buniatyan
Deep Lake: a Lakehouse for Deep Learning
Sep 22, 2022



Abstract:Traditional data lakes provide critical data infrastructure for analytical workloads by enabling time travel, running SQL queries, ingesting data with ACID transactions, and visualizing petabyte-scale datasets on cloud storage. They allow organizations to break down data silos, unlock data-driven decision-making, improve operational efficiency, and reduce costs. However, as deep learning takes over common analytical workflows, traditional data lakes become less useful for applications such as natural language processing (NLP), audio processing, computer vision, and applications involving non-tabular datasets. This paper presents Deep Lake, an open-source lakehouse for deep learning applications developed at Activeloop. Deep Lake maintains the benefits of a vanilla data lake with one key difference: it stores complex data, such as images, videos, annotations, as well as tabular data, in the form of tensors and rapidly streams the data over the network to (a) Tensor Query Language, (b) in-browser visualization engine, or (c) deep learning frameworks without sacrificing GPU utilization. Datasets stored in Deep Lake can be accessed from PyTorch, TensorFlow, JAX, and integrate with numerous MLOps tools.
Hyper: Distributed Cloud Processing for Large-Scale Deep Learning Tasks
Oct 16, 2019



Abstract:Training and deploying deep learning models in real-world applications require processing large amounts of data. This is a challenging task when the amount of data grows to a hundred terabytes, or even, petabyte-scale. We introduce a hybrid distributed cloud framework with a unified view to multiple clouds and an on-premise infrastructure for processing tasks using both CPU and GPU compute instances at scale. The system implements a distributed file system and failure-tolerant task processing scheduler, independent of the language and Deep Learning framework used. It allows to utilize unstable cheap resources on the cloud to significantly reduce costs. We demonstrate the scalability of the framework on running pre-processing, distributed training, hyperparameter search and large-scale inference tasks utilizing 10,000 CPU cores and 300 GPU instances with the overall processing power of 30 petaflops.
Siamese Encoding and Alignment by Multiscale Learning with Self-Supervision
Apr 04, 2019
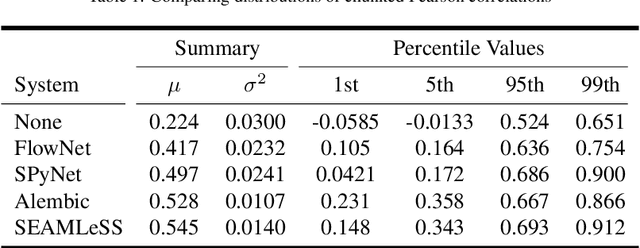
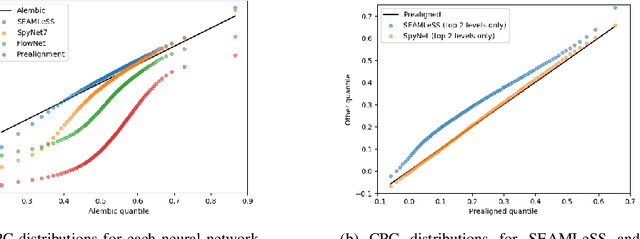

Abstract:We propose a method of aligning a source image to a target image, where the transform is specified by a dense vector field. The two images are encoded as feature hierarchies by siamese convolutional nets. Then a hierarchy of aligner modules computes the transform in a coarse-to-fine recursion. Each module receives as input the transform that was computed by the module at the level above, aligns the source and target encodings at the same level of the hierarchy, and then computes an improved approximation to the transform using a convolutional net. The entire architecture of encoder and aligner nets is trained in a self-supervised manner to minimize the squared error between source and target remaining after alignment. We show that siamese encoding enables more accurate alignment than the image pyramids of SPyNet, a previous deep learning approach to coarse-to-fine alignment. Furthermore, self-supervision applies even without target values for the transform, unlike the strongly supervised SPyNet. We also show that our approach outperforms one-shot approaches to alignment, because the fine pathways in the latter approach may fail to contribute to alignment accuracy when displacements are large. As shown by previous one-shot approaches, good results from self-supervised learning require that the loss function additionally penalize non-smooth transforms. We demonstrate that "masking out" the penalty function near discontinuities leads to correct recovery of non-smooth transforms. Our claims are supported by empirical comparisons using images from serial section electron microscopy of brain tissue.
PZnet: Efficient 3D ConvNet Inference on Manycore CPUs
Mar 18, 2019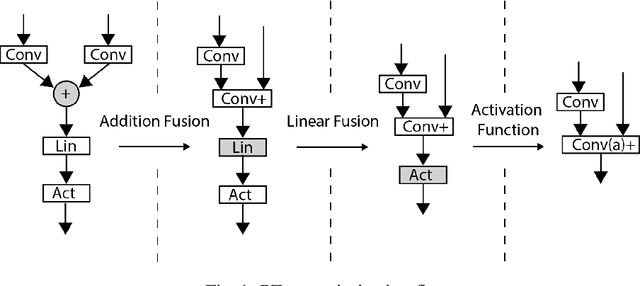



Abstract:Convolutional nets have been shown to achieve state-of-the-art accuracy in many biomedical image analysis tasks. Many tasks within biomedical analysis domain involve analyzing volumetric (3D) data acquired by CT, MRI and Microscopy acquisition methods. To deploy convolutional nets in practical working systems, it is important to solve the efficient inference problem. Namely, one should be able to apply an already-trained convolutional network to many large images using limited computational resources. In this paper we present PZnet, a CPU-only engine that can be used to perform inference for a variety of 3D convolutional net architectures. PZNet outperforms MKL-based CPU implementations of PyTorch and Tensorflow by more than 3.5x for the popular U-net architecture. Moreover, for 3D convolutions with low featuremap numbers, cloud CPU inference with PZnet outperfroms cloud GPU inference in terms of cost efficiency.
Deep Learning Improves Template Matching by Normalized Cross Correlation
May 24, 2017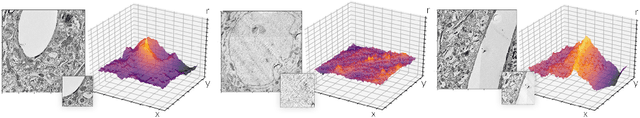

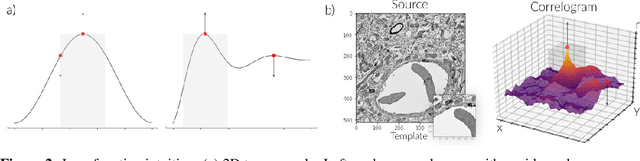
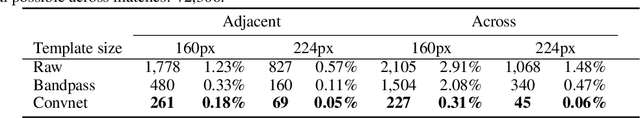
Abstract:Template matching by normalized cross correlation (NCC) is widely used for finding image correspondences. We improve the robustness of this algorithm by preprocessing images with "siamese" convolutional networks trained to maximize the contrast between NCC values of true and false matches. The improvement is quantified using patches of brain images from serial section electron microscopy. Relative to a parameter-tuned bandpass filter, siamese convolutional networks significantly reduce false matches. Furthermore, all false matches can be eliminated by removing a tiny fraction of all matches based on NCC values. The improved accuracy of our method could be essential for connectomics, because emerging petascale datasets may require billions of template matches to assemble 2D images of serial sections into a 3D image stack. Our method is also expected to generalize to many other computer vision applications that use NCC template matching to find image correspondences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge