Thomas Macrina
Automated Neuron Shape Analysis from Electron Microscopy
May 29, 2020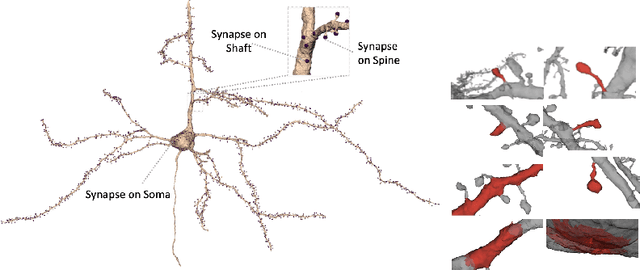
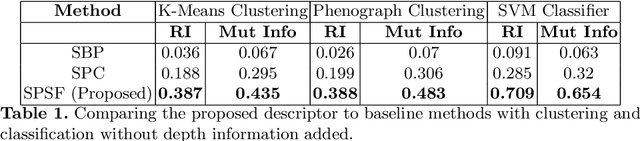
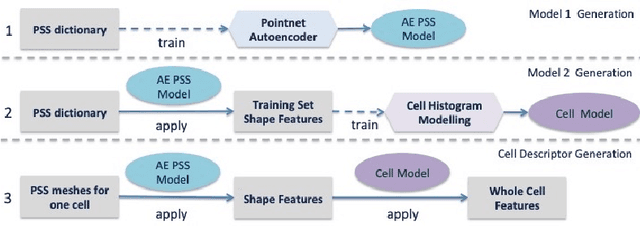
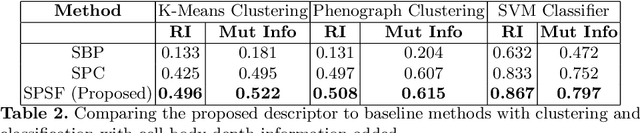
Abstract:Morphology based analysis of cell types has been an area of great interest to the neuroscience community for several decades. Recently, high resolution electron microscopy (EM) datasets of the mouse brain have opened up opportunities for data analysis at a level of detail that was previously impossible. These datasets are very large in nature and thus, manual analysis is not a practical solution. Of particular interest are details to the level of post synaptic structures. This paper proposes a fully automated framework for analysis of post-synaptic structure based neuron analysis from EM data. The processing framework involves shape extraction, representation with an autoencoder, and whole cell modeling and analysis based on shape distributions. We apply our novel framework on a dataset of 1031 neurons obtained from imaging a 1mm x 1mm x 40 micrometer volume of the mouse visual cortex and show the strength of our method in clustering and classification of neuronal shapes.
Convolutional nets for reconstructing neural circuits from brain images acquired by serial section electron microscopy
Apr 29, 2019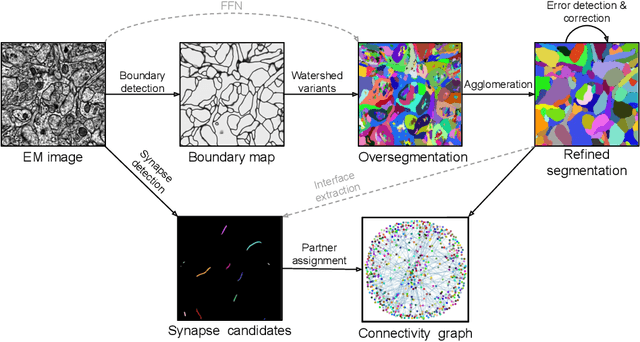
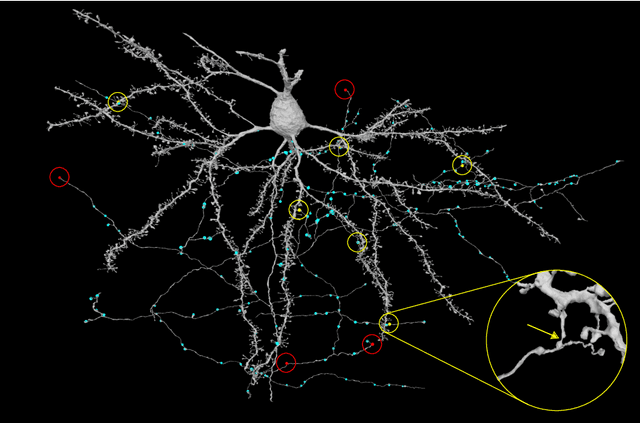

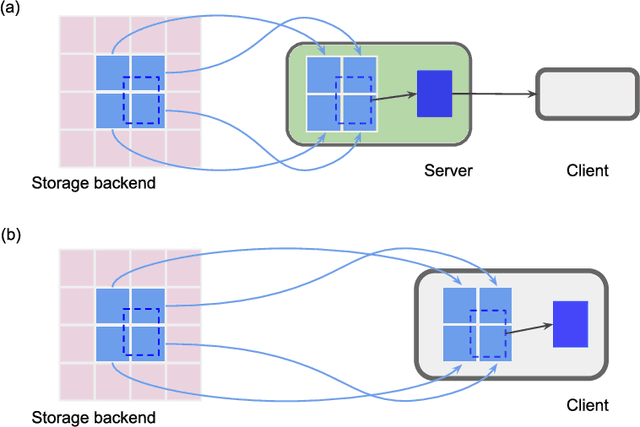
Abstract:Neural circuits can be reconstructed from brain images acquired by serial section electron microscopy. Image analysis has been performed by manual labor for half a century, and efforts at automation date back almost as far. Convolutional nets were first applied to neuronal boundary detection a dozen years ago, and have now achieved impressive accuracy on clean images. Robust handling of image defects is a major outstanding challenge. Convolutional nets are also being employed for other tasks in neural circuit reconstruction: finding synapses and identifying synaptic partners, extending or pruning neuronal reconstructions, and aligning serial section images to create a 3D image stack. Computational systems are being engineered to handle petavoxel images of cubic millimeter brain volumes.
Deep Learning Improves Template Matching by Normalized Cross Correlation
May 24, 2017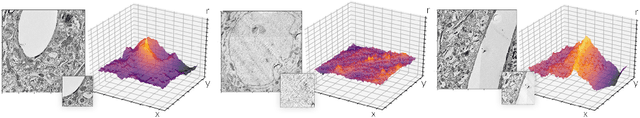

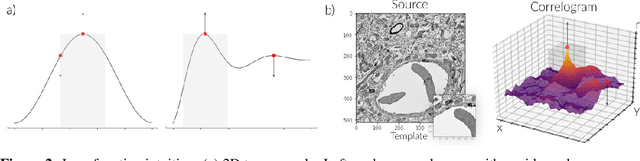
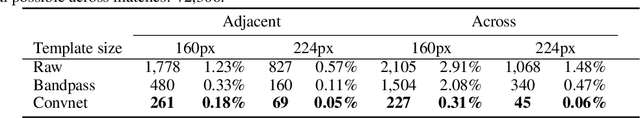
Abstract:Template matching by normalized cross correlation (NCC) is widely used for finding image correspondences. We improve the robustness of this algorithm by preprocessing images with "siamese" convolutional networks trained to maximize the contrast between NCC values of true and false matches. The improvement is quantified using patches of brain images from serial section electron microscopy. Relative to a parameter-tuned bandpass filter, siamese convolutional networks significantly reduce false matches. Furthermore, all false matches can be eliminated by removing a tiny fraction of all matches based on NCC values. The improved accuracy of our method could be essential for connectomics, because emerging petascale datasets may require billions of template matches to assemble 2D images of serial sections into a 3D image stack. Our method is also expected to generalize to many other computer vision applications that use NCC template matching to find image correspondences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge