Kaiming Kuang
Template-Guided Reconstruction of Pulmonary Segments with Neural Implicit Functions
May 13, 2025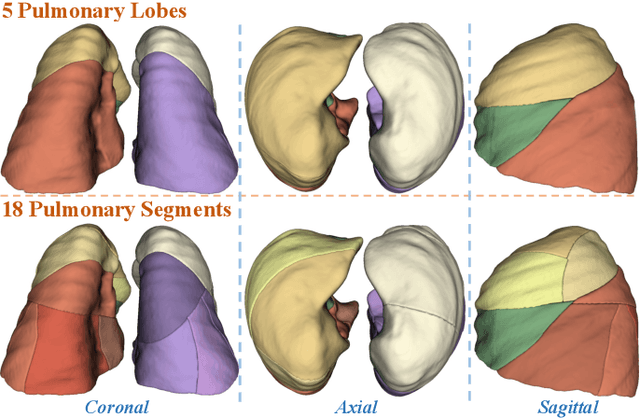
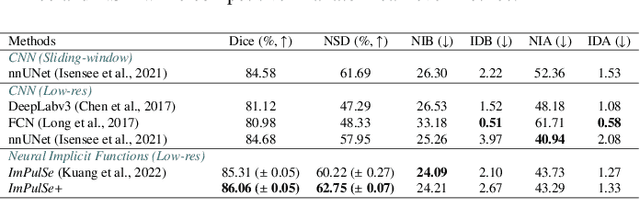
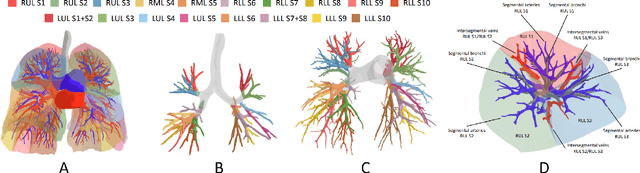
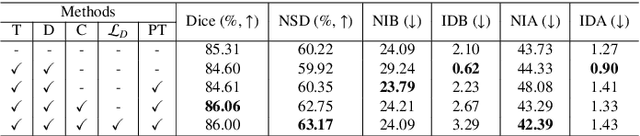
Abstract:High-quality 3D reconstruction of pulmonary segments plays a crucial role in segmentectomy and surgical treatment planning for lung cancer. Due to the resolution requirement of the target reconstruction, conventional deep learning-based methods often suffer from computational resource constraints or limited granularity. Conversely, implicit modeling is favored due to its computational efficiency and continuous representation at any resolution. We propose a neural implicit function-based method to learn a 3D surface to achieve anatomy-aware, precise pulmonary segment reconstruction, represented as a shape by deforming a learnable template. Additionally, we introduce two clinically relevant evaluation metrics to assess the reconstruction comprehensively. Further, due to the absence of publicly available shape datasets to benchmark reconstruction algorithms, we developed a shape dataset named Lung3D, including the 3D models of 800 labeled pulmonary segments and the corresponding airways, arteries, veins, and intersegmental veins. We demonstrate that the proposed approach outperforms existing methods, providing a new perspective for pulmonary segment reconstruction. Code and data will be available at https://github.com/M3DV/ImPulSe.
CyberDemo: Augmenting Simulated Human Demonstration for Real-World Dexterous Manipulation
Mar 01, 2024Abstract:We introduce CyberDemo, a novel approach to robotic imitation learning that leverages simulated human demonstrations for real-world tasks. By incorporating extensive data augmentation in a simulated environment, CyberDemo outperforms traditional in-domain real-world demonstrations when transferred to the real world, handling diverse physical and visual conditions. Regardless of its affordability and convenience in data collection, CyberDemo outperforms baseline methods in terms of success rates across various tasks and exhibits generalizability with previously unseen objects. For example, it can rotate novel tetra-valve and penta-valve, despite human demonstrations only involving tri-valves. Our research demonstrates the significant potential of simulated human demonstrations for real-world dexterous manipulation tasks. More details can be found at https://cyber-demo.github.io
Deep Rib Fracture Instance Segmentation and Classification from CT on the RibFrac Challenge
Feb 14, 2024Abstract:Rib fractures are a common and potentially severe injury that can be challenging and labor-intensive to detect in CT scans. While there have been efforts to address this field, the lack of large-scale annotated datasets and evaluation benchmarks has hindered the development and validation of deep learning algorithms. To address this issue, the RibFrac Challenge was introduced, providing a benchmark dataset of over 5,000 rib fractures from 660 CT scans, with voxel-level instance mask annotations and diagnosis labels for four clinical categories (buckle, nondisplaced, displaced, or segmental). The challenge includes two tracks: a detection (instance segmentation) track evaluated by an FROC-style metric and a classification track evaluated by an F1-style metric. During the MICCAI 2020 challenge period, 243 results were evaluated, and seven teams were invited to participate in the challenge summary. The analysis revealed that several top rib fracture detection solutions achieved performance comparable or even better than human experts. Nevertheless, the current rib fracture classification solutions are hardly clinically applicable, which can be an interesting area in the future. As an active benchmark and research resource, the data and online evaluation of the RibFrac Challenge are available at the challenge website. As an independent contribution, we have also extended our previous internal baseline by incorporating recent advancements in large-scale pretrained networks and point-based rib segmentation techniques. The resulting FracNet+ demonstrates competitive performance in rib fracture detection, which lays a foundation for further research and development in AI-assisted rib fracture detection and diagnosis.
OpenShape: Scaling Up 3D Shape Representation Towards Open-World Understanding
May 18, 2023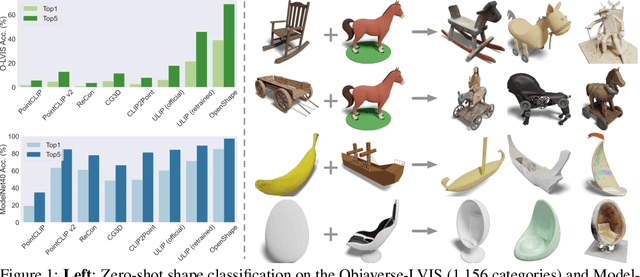
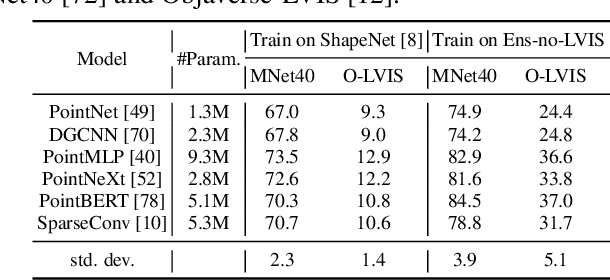
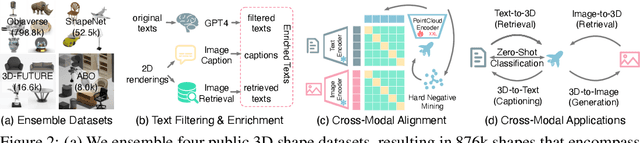
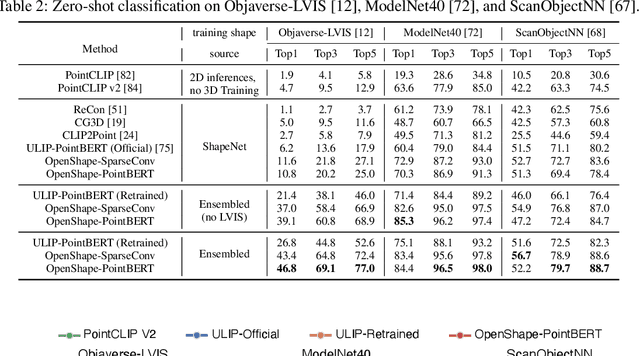
Abstract:We introduce OpenShape, a method for learning multi-modal joint representations of text, image, and point clouds. We adopt the commonly used multi-modal contrastive learning framework for representation alignment, but with a specific focus on scaling up 3D representations to enable open-world 3D shape understanding. To achieve this, we scale up training data by ensembling multiple 3D datasets and propose several strategies to automatically filter and enrich noisy text descriptions. We also explore and compare strategies for scaling 3D backbone networks and introduce a novel hard negative mining module for more efficient training. We evaluate OpenShape on zero-shot 3D classification benchmarks and demonstrate its superior capabilities for open-world recognition. Specifically, OpenShape achieves a zero-shot accuracy of 46.8% on the 1,156-category Objaverse-LVIS benchmark, compared to less than 10% for existing methods. OpenShape also achieves an accuracy of 85.3% on ModelNet40, outperforming previous zero-shot baseline methods by 20% and performing on par with some fully-supervised methods. Furthermore, we show that our learned embeddings encode a wide range of visual and semantic concepts (e.g., subcategories, color, shape, style) and facilitate fine-grained text-3D and image-3D interactions. Due to their alignment with CLIP embeddings, our learned shape representations can also be integrated with off-the-shelf CLIP-based models for various applications, such as point cloud captioning and point cloud-conditioned image generation.
SGDA: Towards 3D Universal Pulmonary Nodule Detection via Slice Grouped Domain Attention
Mar 07, 2023Abstract:Lung cancer is the leading cause of cancer death worldwide. The best solution for lung cancer is to diagnose the pulmonary nodules in the early stage, which is usually accomplished with the aid of thoracic computed tomography (CT). As deep learning thrives, convolutional neural networks (CNNs) have been introduced into pulmonary nodule detection to help doctors in this labor-intensive task and demonstrated to be very effective. However, the current pulmonary nodule detection methods are usually domain-specific, and cannot satisfy the requirement of working in diverse real-world scenarios. To address this issue, we propose a slice grouped domain attention (SGDA) module to enhance the generalization capability of the pulmonary nodule detection networks. This attention module works in the axial, coronal, and sagittal directions. In each direction, we divide the input feature into groups, and for each group, we utilize a universal adapter bank to capture the feature subspaces of the domains spanned by all pulmonary nodule datasets. Then the bank outputs are combined from the perspective of domain to modulate the input group. Extensive experiments demonstrate that SGDA enables substantially better multi-domain pulmonary nodule detection performance compared with the state-of-the-art multi-domain learning methods.
RibSeg v2: A Large-scale Benchmark for Rib Labeling and Anatomical Centerline Extraction
Oct 18, 2022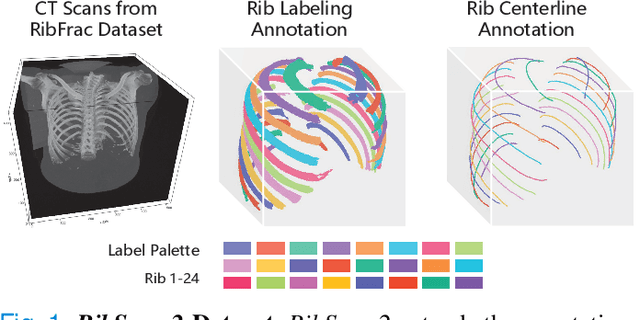
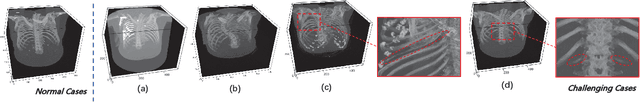
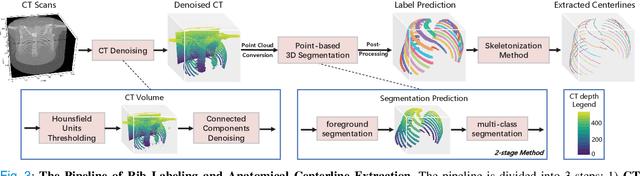
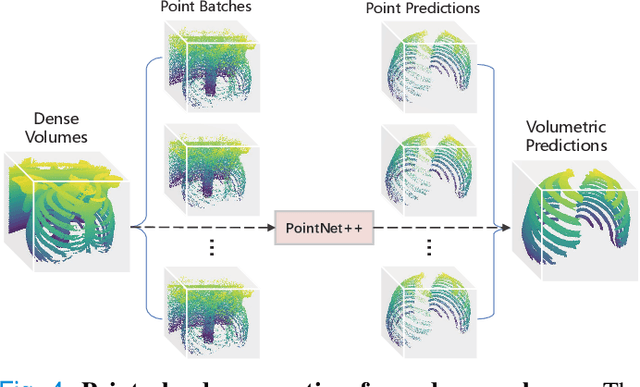
Abstract:Automatic rib labeling and anatomical centerline extraction are common prerequisites for various clinical applications. Prior studies either use in-house datasets that are inaccessible to communities, or focus on rib segmentation that neglects the clinical significance of rib labeling. To address these issues, we extend our prior dataset (RibSeg) on the binary rib segmentation task to a comprehensive benchmark, named RibSeg v2, with 660 CT scans (15,466 individual ribs in total) and annotations manually inspected by experts for rib labeling and anatomical centerline extraction. Based on the RibSeg v2, we develop a pipeline including deep learning-based methods for rib labeling, and a skeletonization-based method for centerline extraction. To improve computational efficiency, we propose a sparse point cloud representation of CT scans and compare it with standard dense voxel grids. Moreover, we design and analyze evaluation metrics to address the key challenges of each task. Our dataset, code, and model are available online to facilitate open research at https://github.com/M3DV/RibSeg
LSSANet: A Long Short Slice-Aware Network for Pulmonary Nodule Detection
Aug 03, 2022



Abstract:Convolutional neural networks (CNNs) have been demonstrated to be highly effective in the field of pulmonary nodule detection. However, existing CNN based pulmonary nodule detection methods lack the ability to capture long-range dependencies, which is vital for global information extraction. In computer vision tasks, non-local operations have been widely utilized, but the computational cost could be very high for 3D computed tomography (CT) images. To address this issue, we propose a long short slice-aware network (LSSANet) for the detection of pulmonary nodules. In particular, we develop a new non-local mechanism termed long short slice grouping (LSSG), which splits the compact non-local embeddings into a short-distance slice grouped one and a long-distance slice grouped counterpart. This not only reduces the computational burden, but also keeps long-range dependencies among any elements across slices and in the whole feature map. The proposed LSSG is easy-to-use and can be plugged into many pulmonary nodule detection networks. To verify the performance of LSSANet, we compare with several recently proposed and competitive detection approaches based on 2D/3D CNN. Promising evaluation results on the large-scale PN9 dataset demonstrate the effectiveness of our method. Code is at https://github.com/Ruixxxx/LSSANet.
What Makes for Automatic Reconstruction of Pulmonary Segments
Jul 14, 2022



Abstract:3D reconstruction of pulmonary segments plays an important role in surgical treatment planning of lung cancer, which facilitates preservation of pulmonary function and helps ensure low recurrence rates. However, automatic reconstruction of pulmonary segments remains unexplored in the era of deep learning. In this paper, we investigate what makes for automatic reconstruction of pulmonary segments. First and foremost, we formulate, clinically and geometrically, the anatomical definitions of pulmonary segments, and propose evaluation metrics adhering to these definitions. Second, we propose ImPulSe (Implicit Pulmonary Segment), a deep implicit surface model designed for pulmonary segment reconstruction. The automatic reconstruction of pulmonary segments by ImPulSe is accurate in metrics and visually appealing. Compared with canonical segmentation methods, ImPulSe outputs continuous predictions of arbitrary resolutions with higher training efficiency and fewer parameters. Lastly, we experiment with different network inputs to analyze what matters in the task of pulmonary segment reconstruction. Our code is available at https://github.com/M3DV/ImPulSe.
Asymmetric 3D Context Fusion for Universal Lesion Detection
Sep 17, 2021



Abstract:Modeling 3D context is essential for high-performance 3D medical image analysis. Although 2D networks benefit from large-scale 2D supervised pretraining, it is weak in capturing 3D context. 3D networks are strong in 3D context yet lack supervised pretraining. As an emerging technique, \emph{3D context fusion operator}, which enables conversion from 2D pretrained networks, leverages the advantages of both and has achieved great success. Existing 3D context fusion operators are designed to be spatially symmetric, i.e., performing identical operations on each 2D slice like convolutions. However, these operators are not truly equivariant to translation, especially when only a few 3D slices are used as inputs. In this paper, we propose a novel asymmetric 3D context fusion operator (A3D), which uses different weights to fuse 3D context from different 2D slices. Notably, A3D is NOT translation-equivariant while it significantly outperforms existing symmetric context fusion operators without introducing large computational overhead. We validate the effectiveness of the proposed method by extensive experiments on DeepLesion benchmark, a large-scale public dataset for universal lesion detection from computed tomography (CT). The proposed A3D consistently outperforms symmetric context fusion operators by considerable margins, and establishes a new \emph{state of the art} on DeepLesion. To facilitate open research, our code and model in PyTorch are available at https://github.com/M3DV/AlignShift.
MIA-Prognosis: A Deep Learning Framework to Predict Therapy Response
Oct 08, 2020


Abstract:Predicting clinical outcome is remarkably important but challenging. Research efforts have been paid on seeking significant biomarkers associated with the therapy response or/and patient survival. However, these biomarkers are generally costly and invasive, and possibly dissatifactory for novel therapy. On the other hand, multi-modal, heterogeneous, unaligned temporal data is continuously generated in clinical practice. This paper aims at a unified deep learning approach to predict patient prognosis and therapy response, with easily accessible data, e.g., radiographics, laboratory and clinical information. Prior arts focus on modeling single data modality, or ignore the temporal changes. Importantly, the clinical time series is asynchronous in practice, i.e., recorded with irregular intervals. In this study, we formalize the prognosis modeling as a multi-modal asynchronous time series classification task, and propose a MIA-Prognosis framework with Measurement, Intervention and Assessment (MIA) information to predict therapy response, where a Simple Temporal Attention (SimTA) module is developed to process the asynchronous time series. Experiments on synthetic dataset validate the superiory of SimTA over standard RNN-based approaches. Furthermore, we experiment the proposed method on an in-house, retrospective dataset of real-world non-small cell lung cancer patients under anti-PD-1 immunotherapy. The proposed method achieves promising performance on predicting the immunotherapy response. Notably, our predictive model could further stratify low-risk and high-risk patients in terms of long-term survival.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge