Shizhong Han
Generative Scenario Rollouts for End-to-End Autonomous Driving
Jan 16, 2026Abstract:Vision-Language-Action (VLA) models are emerging as highly effective planning models for end-to-end autonomous driving systems. However, current works mostly rely on imitation learning from sparse trajectory annotations and under-utilize their potential as generative models. We propose Generative Scenario Rollouts (GeRo), a plug-and-play framework for VLA models that jointly performs planning and generation of language-grounded future traffic scenes through an autoregressive rollout strategy. First, a VLA model is trained to encode ego vehicle and agent dynamics into latent tokens under supervision from planning, motion, and language tasks, facilitating text-aligned generation. Next, GeRo performs language-conditioned autoregressive generation. Given multi-view images, a scenario description, and ego-action questions, it generates future latent tokens and textual responses to guide long-horizon rollouts. A rollout-consistency loss stabilizes predictions using ground truth or pseudo-labels, mitigating drift and preserving text-action alignment. This design enables GeRo to perform temporally consistent, language-grounded rollouts that support long-horizon reasoning and multi-agent planning. On Bench2Drive, GeRo improves driving score and success rate by +15.7 and +26.2, respectively. By integrating reinforcement learning with generative rollouts, GeRo achieves state-of-the-art closed-loop and open-loop performance, demonstrating strong zero-shot robustness. These results highlight the promise of generative, language-conditioned reasoning as a foundation for safer and more interpretable end-to-end autonomous driving.
ODG: Occupancy Prediction Using Dual Gaussians
Jun 12, 2025Abstract:Occupancy prediction infers fine-grained 3D geometry and semantics from camera images of the surrounding environment, making it a critical perception task for autonomous driving. Existing methods either adopt dense grids as scene representation, which is difficult to scale to high resolution, or learn the entire scene using a single set of sparse queries, which is insufficient to handle the various object characteristics. In this paper, we present ODG, a hierarchical dual sparse Gaussian representation to effectively capture complex scene dynamics. Building upon the observation that driving scenes can be universally decomposed into static and dynamic counterparts, we define dual Gaussian queries to better model the diverse scene objects. We utilize a hierarchical Gaussian transformer to predict the occupied voxel centers and semantic classes along with the Gaussian parameters. Leveraging the real-time rendering capability of 3D Gaussian Splatting, we also impose rendering supervision with available depth and semantic map annotations injecting pixel-level alignment to boost occupancy learning. Extensive experiments on the Occ3D-nuScenes and Occ3D-Waymo benchmarks demonstrate our proposed method sets new state-of-the-art results while maintaining low inference cost.
RoCA: Robust Cross-Domain End-to-End Autonomous Driving
Jun 11, 2025Abstract:End-to-end (E2E) autonomous driving has recently emerged as a new paradigm, offering significant potential. However, few studies have looked into the practical challenge of deployment across domains (e.g., cities). Although several works have incorporated Large Language Models (LLMs) to leverage their open-world knowledge, LLMs do not guarantee cross-domain driving performance and may incur prohibitive retraining costs during domain adaptation. In this paper, we propose RoCA, a novel framework for robust cross-domain E2E autonomous driving. RoCA formulates the joint probabilistic distribution over the tokens that encode ego and surrounding vehicle information in the E2E pipeline. Instantiating with a Gaussian process (GP), RoCA learns a set of basis tokens with corresponding trajectories, which span diverse driving scenarios. Then, given any driving scene, it is able to probabilistically infer the future trajectory. By using RoCA together with a base E2E model in source-domain training, we improve the generalizability of the base model, without requiring extra inference computation. In addition, RoCA enables robust adaptation on new target domains, significantly outperforming direct finetuning. We extensively evaluate RoCA on various cross-domain scenarios and show that it achieves strong domain generalization and adaptation performance.
DySS: Dynamic Queries and State-Space Learning for Efficient 3D Object Detection from Multi-Camera Videos
Jun 11, 2025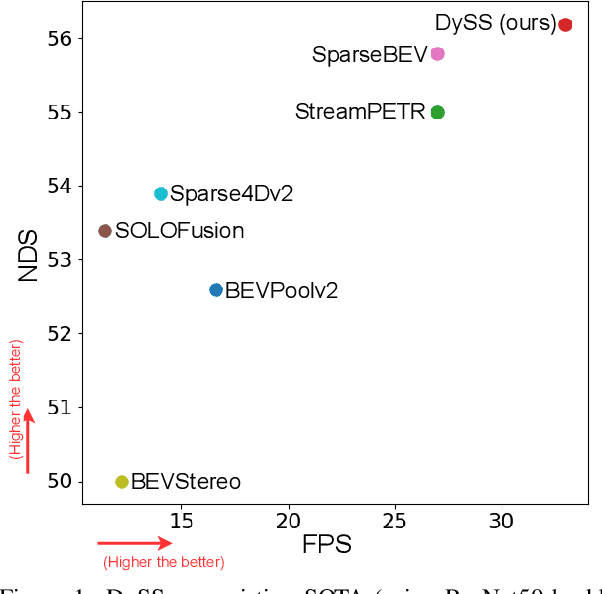
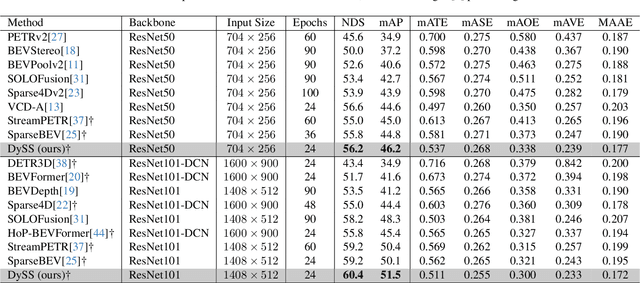
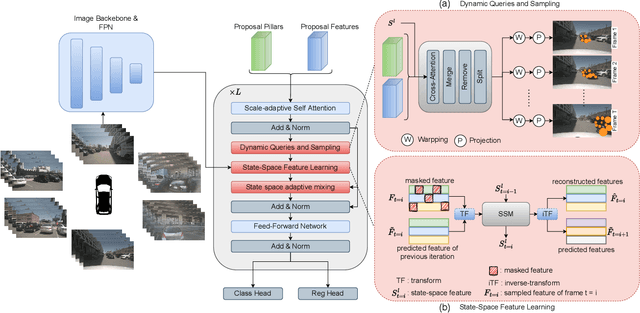
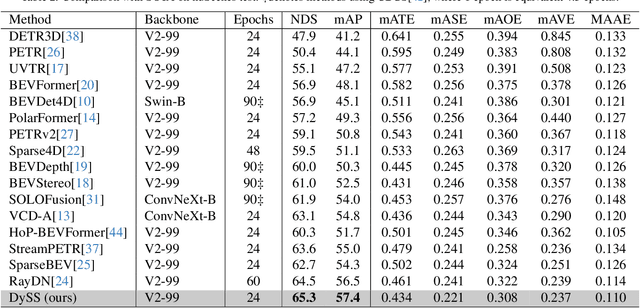
Abstract:Camera-based 3D object detection in Bird's Eye View (BEV) is one of the most important perception tasks in autonomous driving. Earlier methods rely on dense BEV features, which are costly to construct. More recent works explore sparse query-based detection. However, they still require a large number of queries and can become expensive to run when more video frames are used. In this paper, we propose DySS, a novel method that employs state-space learning and dynamic queries. More specifically, DySS leverages a state-space model (SSM) to sequentially process the sampled features over time steps. In order to encourage the model to better capture the underlying motion and correspondence information, we introduce auxiliary tasks of future prediction and masked reconstruction to better train the SSM. The state of the SSM then provides an informative yet efficient summarization of the scene. Based on the state-space learned features, we dynamically update the queries via merge, remove, and split operations, which help maintain a useful, lean set of detection queries throughout the network. Our proposed DySS achieves both superior detection performance and efficient inference. Specifically, on the nuScenes test split, DySS achieves 65.31 NDS and 57.4 mAP, outperforming the latest state of the art. On the val split, DySS achieves 56.2 NDS and 46.2 mAP, as well as a real-time inference speed of 33 FPS.
BePo: Leveraging Birds Eye View and Sparse Points for Efficient and Accurate 3D Occupancy Prediction
Jun 08, 2025Abstract:3D occupancy provides fine-grained 3D geometry and semantics for scene understanding which is critical for autonomous driving. Most existing methods, however, carry high compute costs, requiring dense 3D feature volume and cross-attention to effectively aggregate information. More recent works have adopted Bird's Eye View (BEV) or sparse points as scene representation with much reduced cost, but still suffer from their respective shortcomings. More concretely, BEV struggles with small objects that often experience significant information loss after being projected to the ground plane. On the other hand, points can flexibly model little objects in 3D, but is inefficient at capturing flat surfaces or large objects. To address these challenges, in this paper, we present a novel 3D occupancy prediction approach, BePo, which combines BEV and sparse points based representations. We propose a dual-branch design: a query-based sparse points branch and a BEV branch. The 3D information learned in the sparse points branch is shared with the BEV stream via cross-attention, which enriches the weakened signals of difficult objects on the BEV plane. The outputs of both branches are finally fused to generate predicted 3D occupancy. We conduct extensive experiments on the Occ3D-nuScenes and Occ3D-Waymo benchmarks that demonstrate the superiority of our proposed BePo. Moreover, BePo also delivers competitive inference speed when compared to the latest efficient approaches.
FALO: Fast and Accurate LiDAR 3D Object Detection on Resource-Constrained Devices
Jun 04, 2025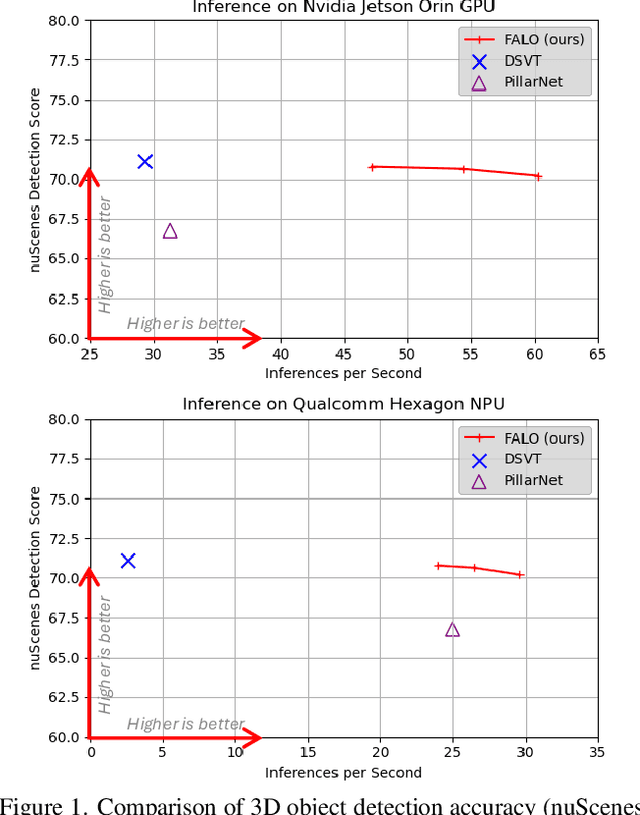
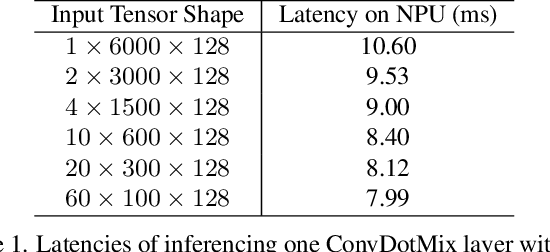
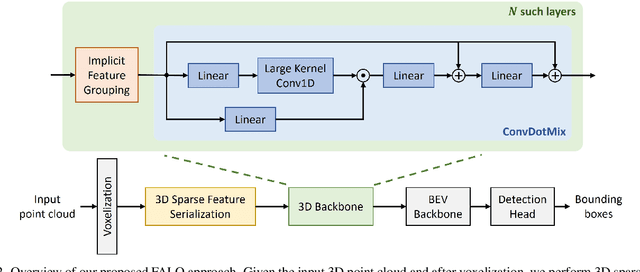
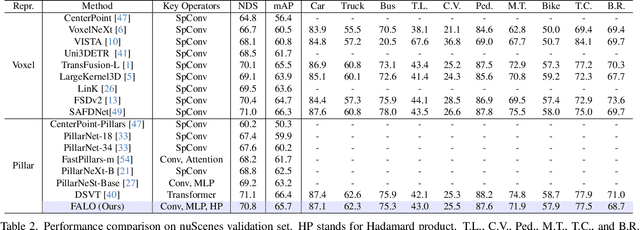
Abstract:Existing LiDAR 3D object detection methods predominantely rely on sparse convolutions and/or transformers, which can be challenging to run on resource-constrained edge devices, due to irregular memory access patterns and high computational costs. In this paper, we propose FALO, a hardware-friendly approach to LiDAR 3D detection, which offers both state-of-the-art (SOTA) detection accuracy and fast inference speed. More specifically, given the 3D point cloud and after voxelization, FALO first arranges sparse 3D voxels into a 1D sequence based on their coordinates and proximity. The sequence is then processed by our proposed ConvDotMix blocks, consisting of large-kernel convolutions, Hadamard products, and linear layers. ConvDotMix provides sufficient mixing capability in both spatial and embedding dimensions, and introduces higher-order nonlinear interaction among spatial features. Furthermore, when going through the ConvDotMix layers, we introduce implicit grouping, which balances the tensor dimensions for more efficient inference and takes into account the growing receptive field. All these operations are friendly to run on resource-constrained platforms and proposed FALO can readily deploy on compact, embedded devices. Our extensive evaluation on LiDAR 3D detection benchmarks such as nuScenes and Waymo shows that FALO achieves competitive performance. Meanwhile, FALO is 1.6~9.8x faster than the latest SOTA on mobile Graphics Processing Unit (GPU) and mobile Neural Processing Unit (NPU).
Distilling Multi-modal Large Language Models for Autonomous Driving
Jan 16, 2025
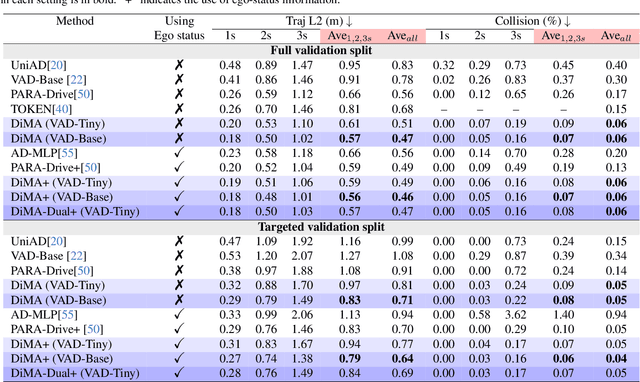

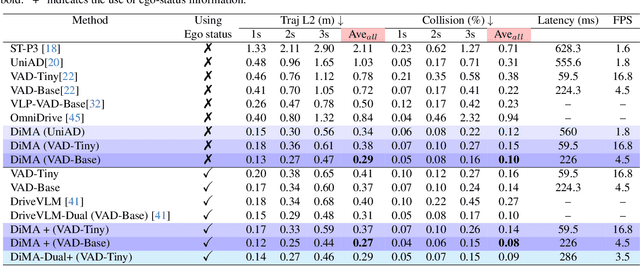
Abstract:Autonomous driving demands safe motion planning, especially in critical "long-tail" scenarios. Recent end-to-end autonomous driving systems leverage large language models (LLMs) as planners to improve generalizability to rare events. However, using LLMs at test time introduces high computational costs. To address this, we propose DiMA, an end-to-end autonomous driving system that maintains the efficiency of an LLM-free (or vision-based) planner while leveraging the world knowledge of an LLM. DiMA distills the information from a multi-modal LLM to a vision-based end-to-end planner through a set of specially designed surrogate tasks. Under a joint training strategy, a scene encoder common to both networks produces structured representations that are semantically grounded as well as aligned to the final planning objective. Notably, the LLM is optional at inference, enabling robust planning without compromising on efficiency. Training with DiMA results in a 37% reduction in the L2 trajectory error and an 80% reduction in the collision rate of the vision-based planner, as well as a 44% trajectory error reduction in longtail scenarios. DiMA also achieves state-of-the-art performance on the nuScenes planning benchmark.
Ranking and Combining Latent Structured Predictive Scores without Labeled Data
Aug 14, 2024


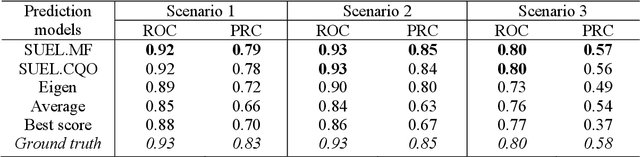
Abstract:Combining multiple predictors obtained from distributed data sources to an accurate meta-learner is promising to achieve enhanced performance in lots of prediction problems. As the accuracy of each predictor is usually unknown, integrating the predictors to achieve better performance is challenging. Conventional ensemble learning methods assess the accuracy of predictors based on extensive labeled data. In practical applications, however, the acquisition of such labeled data can prove to be an arduous task. Furthermore, the predictors under consideration may exhibit high degrees of correlation, particularly when similar data sources or machine learning algorithms were employed during their model training. In response to these challenges, this paper introduces a novel structured unsupervised ensemble learning model (SUEL) to exploit the dependency between a set of predictors with continuous predictive scores, rank the predictors without labeled data and combine them to an ensembled score with weights. Two novel correlation-based decomposition algorithms are further proposed to estimate the SUEL model, constrained quadratic optimization (SUEL.CQO) and matrix-factorization-based (SUEL.MF) approaches. The efficacy of the proposed methods is rigorously assessed through both simulation studies and real-world application of risk genes discovery. The results compellingly demonstrate that the proposed methods can efficiently integrate the dependent predictors to an ensemble model without the need of ground truth data.
PADRe: A Unifying Polynomial Attention Drop-in Replacement for Efficient Vision Transformer
Jul 16, 2024



Abstract:We present Polynomial Attention Drop-in Replacement (PADRe), a novel and unifying framework designed to replace the conventional self-attention mechanism in transformer models. Notably, several recent alternative attention mechanisms, including Hyena, Mamba, SimA, Conv2Former, and Castling-ViT, can be viewed as specific instances of our PADRe framework. PADRe leverages polynomial functions and draws upon established results from approximation theory, enhancing computational efficiency without compromising accuracy. PADRe's key components include multiplicative nonlinearities, which we implement using straightforward, hardware-friendly operations such as Hadamard products, incurring only linear computational and memory costs. PADRe further avoids the need for using complex functions such as Softmax, yet it maintains comparable or superior accuracy compared to traditional self-attention. We assess the effectiveness of PADRe as a drop-in replacement for self-attention across diverse computer vision tasks. These tasks include image classification, image-based 2D object detection, and 3D point cloud object detection. Empirical results demonstrate that PADRe runs significantly faster than the conventional self-attention (11x ~ 43x faster on server GPU and mobile NPU) while maintaining similar accuracy when substituting self-attention in the transformer models.
FutureDepth: Learning to Predict the Future Improves Video Depth Estimation
Mar 19, 2024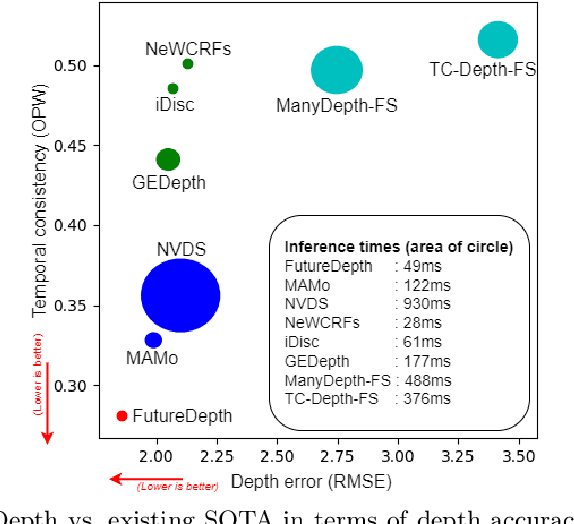
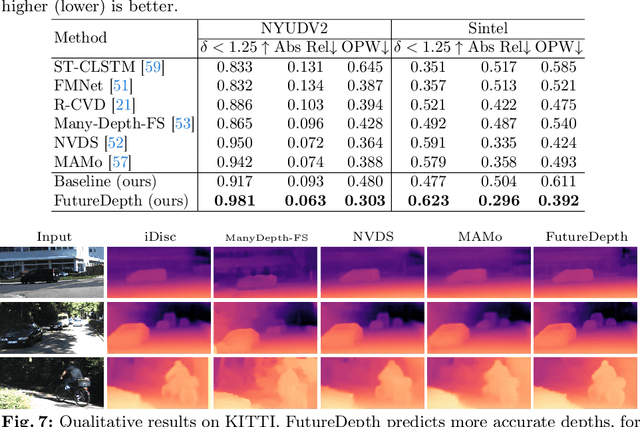
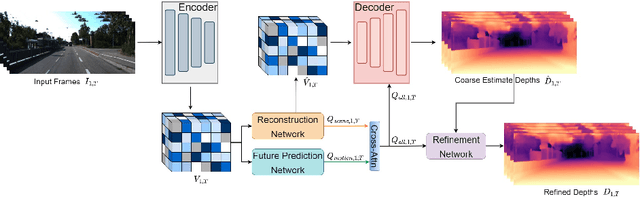
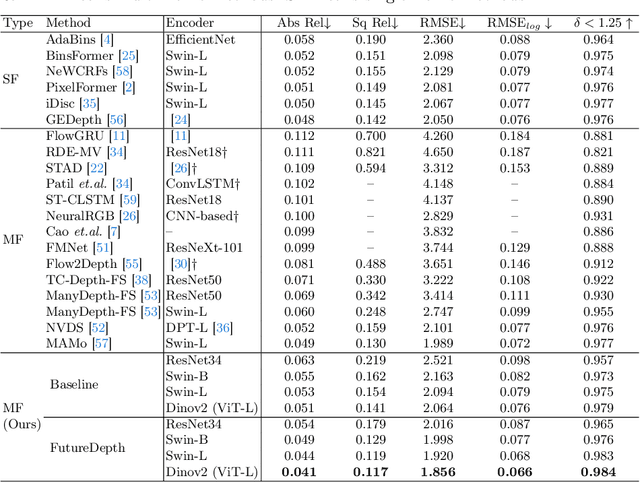
Abstract:In this paper, we propose a novel video depth estimation approach, FutureDepth, which enables the model to implicitly leverage multi-frame and motion cues to improve depth estimation by making it learn to predict the future at training. More specifically, we propose a future prediction network, F-Net, which takes the features of multiple consecutive frames and is trained to predict multi-frame features one time step ahead iteratively. In this way, F-Net learns the underlying motion and correspondence information, and we incorporate its features into the depth decoding process. Additionally, to enrich the learning of multiframe correspondence cues, we further leverage a reconstruction network, R-Net, which is trained via adaptively masked auto-encoding of multiframe feature volumes. At inference time, both F-Net and R-Net are used to produce queries to work with the depth decoder, as well as a final refinement network. Through extensive experiments on several benchmarks, i.e., NYUDv2, KITTI, DDAD, and Sintel, which cover indoor, driving, and open-domain scenarios, we show that FutureDepth significantly improves upon baseline models, outperforms existing video depth estimation methods, and sets new state-of-the-art (SOTA) accuracy. Furthermore, FutureDepth is more efficient than existing SOTA video depth estimation models and has similar latencies when comparing to monocular models
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge