Zhizhong Chai
Deep Rib Fracture Instance Segmentation and Classification from CT on the RibFrac Challenge
Feb 14, 2024Abstract:Rib fractures are a common and potentially severe injury that can be challenging and labor-intensive to detect in CT scans. While there have been efforts to address this field, the lack of large-scale annotated datasets and evaluation benchmarks has hindered the development and validation of deep learning algorithms. To address this issue, the RibFrac Challenge was introduced, providing a benchmark dataset of over 5,000 rib fractures from 660 CT scans, with voxel-level instance mask annotations and diagnosis labels for four clinical categories (buckle, nondisplaced, displaced, or segmental). The challenge includes two tracks: a detection (instance segmentation) track evaluated by an FROC-style metric and a classification track evaluated by an F1-style metric. During the MICCAI 2020 challenge period, 243 results were evaluated, and seven teams were invited to participate in the challenge summary. The analysis revealed that several top rib fracture detection solutions achieved performance comparable or even better than human experts. Nevertheless, the current rib fracture classification solutions are hardly clinically applicable, which can be an interesting area in the future. As an active benchmark and research resource, the data and online evaluation of the RibFrac Challenge are available at the challenge website. As an independent contribution, we have also extended our previous internal baseline by incorporating recent advancements in large-scale pretrained networks and point-based rib segmentation techniques. The resulting FracNet+ demonstrates competitive performance in rib fracture detection, which lays a foundation for further research and development in AI-assisted rib fracture detection and diagnosis.
Deep Omni-supervised Learning for Rib Fracture Detection from Chest Radiology Images
Jun 23, 2023



Abstract:Deep learning (DL)-based rib fracture detection has shown promise of playing an important role in preventing mortality and improving patient outcome. Normally, developing DL-based object detection models requires huge amount of bounding box annotation. However, annotating medical data is time-consuming and expertise-demanding, making obtaining a large amount of fine-grained annotations extremely infeasible. This poses pressing need of developing label-efficient detection models to alleviate radiologists' labeling burden. To tackle this challenge, the literature of object detection has witnessed an increase of weakly-supervised and semi-supervised approaches, yet still lacks a unified framework that leverages various forms of fully-labeled, weakly-labeled, and unlabeled data. In this paper, we present a novel omni-supervised object detection network, ORF-Netv2, to leverage as much available supervision as possible. Specifically, a multi-branch omni-supervised detection head is introduced with each branch trained with a specific type of supervision. A co-training-based dynamic label assignment strategy is then proposed to enable flexibly and robustly learning from the weakly-labeled and unlabeled data. Extensively evaluation was conducted for the proposed framework with three rib fracture datasets on both chest CT and X-ray. By leveraging all forms of supervision, ORF-Netv2 achieves mAPs of 34.7, 44.7, and 19.4 on the three datasets, respectively, surpassing the baseline detector which uses only box annotations by mAP gains of 3.8, 4.8, and 5.0, respectively. Furthermore, ORF-Netv2 consistently outperforms other competitive label-efficient methods over various scenarios, showing a promising framework for label-efficient fracture detection.
ORF-Net: Deep Omni-supervised Rib Fracture Detection from Chest CT Scans
Jul 05, 2022


Abstract:Most of the existing object detection works are based on the bounding box annotation: each object has a precise annotated box. However, for rib fractures, the bounding box annotation is very labor-intensive and time-consuming because radiologists need to investigate and annotate the rib fractures on a slice-by-slice basis. Although a few studies have proposed weakly-supervised methods or semi-supervised methods, they could not handle different forms of supervision simultaneously. In this paper, we proposed a novel omni-supervised object detection network, which can exploit multiple different forms of annotated data to further improve the detection performance. Specifically, the proposed network contains an omni-supervised detection head, in which each form of annotation data corresponds to a unique classification branch. Furthermore, we proposed a dynamic label assignment strategy for different annotated forms of data to facilitate better learning for each branch. Moreover, we also design a confidence-aware classification loss to emphasize the samples with high confidence and further improve the model's performance. Extensive experiments conducted on the testing dataset show our proposed method outperforms other state-of-the-art approaches consistently, demonstrating the efficacy of deep omni-supervised learning on improving rib fracture detection performance.
Conquering Data Variations in Resolution: A Slice-Aware Multi-Branch Decoder Network
Mar 07, 2022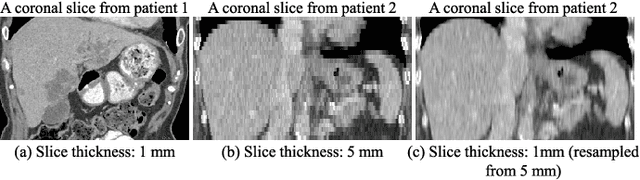
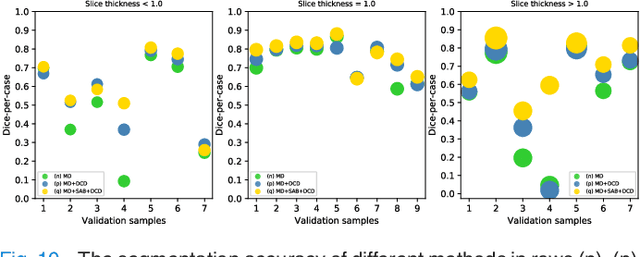
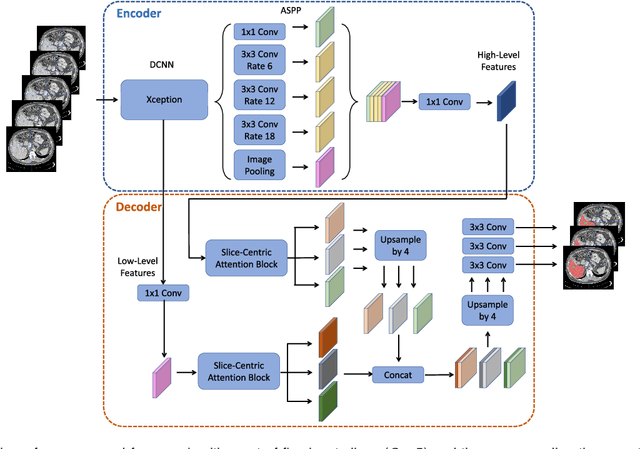
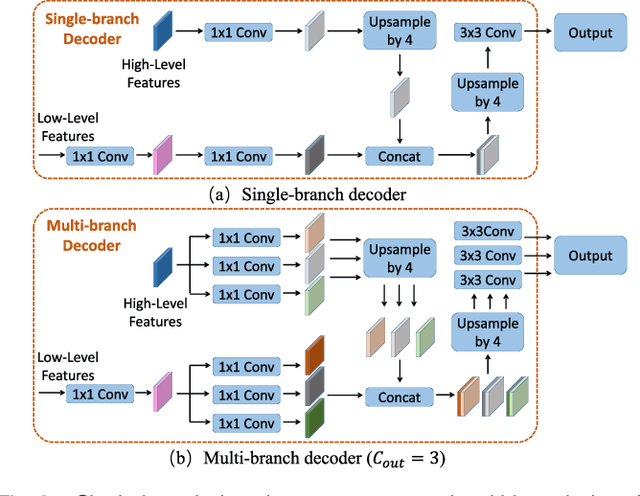
Abstract:Fully convolutional neural networks have made promising progress in joint liver and liver tumor segmentation. Instead of following the debates over 2D versus 3D networks (for example, pursuing the balance between large-scale 2D pretraining and 3D context), in this paper, we novelly identify the wide variation in the ratio between intra- and inter-slice resolutions as a crucial obstacle to the performance. To tackle the mismatch between the intra- and inter-slice information, we propose a slice-aware 2.5D network that emphasizes extracting discriminative features utilizing not only in-plane semantics but also out-of-plane coherence for each separate slice. Specifically, we present a slice-wise multi-input multi-output architecture to instantiate such a design paradigm, which contains a Multi-Branch Decoder (MD) with a Slice-centric Attention Block (SAB) for learning slice-specific features and a Densely Connected Dice (DCD) loss to regularize the inter-slice predictions to be coherent and continuous. Based on the aforementioned innovations, we achieve state-of-the-art results on the MICCAI 2017 Liver Tumor Segmentation (LiTS) dataset. Besides, we also test our model on the ISBI 2019 Segmentation of THoracic Organs at Risk (SegTHOR) dataset, and the result proves the robustness and generalizability of the proposed method in other segmentation tasks.
Deep Semi-supervised Metric Learning with Dual Alignment for Cervical Cancer Cell Detection
Apr 07, 2021

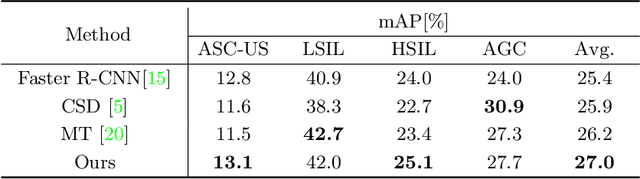

Abstract:With availability of huge amounts of labeled data, deep learning has achieved unprecedented success in various object detection tasks. However, large-scale annotations for medical images are extremely challenging to be acquired due to the high demand of labour and expertise. To address this difficult issue, in this paper we propose a novel semi-supervised deep metric learning method to effectively leverage both labeled and unlabeled data with application to cervical cancer cell detection. Different from previous methods, our model learns an embedding metric space and conducts dual alignment of semantic features on both the proposal and prototype levels. First, on the proposal level, we generate pseudo labels for the unlabeled data to align the proposal features with learnable class proxies derived from the labeled data. Furthermore, we align the prototypes generated from each mini-batch of labeled and unlabeled data to alleviate the influence of possibly noisy pseudo labels. Moreover, we adopt a memory bank to store the labeled prototypes and hence significantly enrich the metric learning information from larger batches. To comprehensively validate the method, we construct a large-scale dataset for semi-supervised cervical cancer cell detection for the first time, consisting of 240,860 cervical cell images in total. Extensive experiments show our proposed method outperforms other state-of-the-art semi-supervised approaches consistently, demonstrating efficacy of deep semi-supervised metric learning with dual alignment on improving cervical cancer cell detection performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge