Dong Wei
Visible Light Positioning With Lamé Curve LEDs: A Generic Approach for Camera Pose Estimation
Feb 02, 2026Abstract:Camera-based visible light positioning (VLP) is a promising technique for accurate and low-cost indoor camera pose estimation (CPE). To reduce the number of required light-emitting diodes (LEDs), advanced methods commonly exploit LED shape features for positioning. Although interesting, they are typically restricted to a single LED geometry, leading to failure in heterogeneous LED-shape scenarios. To address this challenge, this paper investigates Lamé curves as a unified representation of common LED shapes and proposes a generic VLP algorithm using Lamé curve-shaped LEDs, termed LC-VLP. In the considered system, multiple ceiling-mounted Lamé curve-shaped LEDs periodically broadcast their curve parameters via visible light communication, which are captured by a camera-equipped receiver. Based on the received LED images and curve parameters, the receiver can estimate the camera pose using LC-VLP. Specifically, an LED database is constructed offline to store the curve parameters, while online positioning is formulated as a nonlinear least-squares problem and solved iteratively. To provide a reliable initialization, a correspondence-free perspective-\textit{n}-points (FreeP\textit{n}P) algorithm is further developed, enabling approximate CPE without any pre-calibrated reference points. The performance of LC-VLP is verified by both simulations and experiments. Simulations show that LC-VLP outperforms state-of-the-art methods in both circular- and rectangular-LED scenarios, achieving reductions of over 40% in position error and 25% in rotation error. Experiments further show that LC-VLP can achieve an average position accuracy of less than 4 cm.
Division-based Receiver-agnostic RFF Identification in WiFi Systems
Nov 19, 2025Abstract:In physical-layer security schemes, radio frequency fingerprint (RFF) identification of WiFi devices is susceptible to receiver differences, which can significantly degrade classification performance when a model is trained on one receiver but tested on another. In this paper, we propose a division-based receiver-agnostic RFF extraction method for WiFi systems, which removes the receivers' effects by dividing different preambles in the frequency domain. The proposed method requires only a single receiver for training and does not rely on additional calibration or stacking processes. First, for flat fading channel scenarios, the legacy short training field (L-STF) and legacy long training field (L-LTF) of the unknown device are divided by those of the reference device in the frequency domain. The receiver-dependent effects can be eliminated with the requirement of only a single receiver for training, and the higher-dimensional RFF features can be extracted. Second, for frequency-selective fading channel scenarios, the high-throughput long training field (HT-LTF) is divided by the L-LTF in the frequency domain. Only a single receiver is required for training and the higher-dimensional RFF features that are both channel-invariant and receiver-agnostic are extracted. Finally, simulation and experimental results demonstrate that the proposed method effectively mitigate the impacts of channel variations and receiver differences. The classification results show that, even when training on a single receiver and testing on a different one, the proposed method achieves classification accuracy improvements of 15.5% and 28.45% over the state-of-the-art approach in flat fading and frequency-selective fading channel scenarios, respectively.
The Butterfly Effect in Pathology: Exploring Security in Pathology Foundation Models
May 30, 2025Abstract:With the widespread adoption of pathology foundation models in both research and clinical decision support systems, exploring their security has become a critical concern. However, despite their growing impact, the vulnerability of these models to adversarial attacks remains largely unexplored. In this work, we present the first systematic investigation into the security of pathology foundation models for whole slide image~(WSI) analysis against adversarial attacks. Specifically, we introduce the principle of \textit{local perturbation with global impact} and propose a label-free attack framework that operates without requiring access to downstream task labels. Under this attack framework, we revise four classical white-box attack methods and redefine the perturbation budget based on the characteristics of WSI. We conduct comprehensive experiments on three representative pathology foundation models across five datasets and six downstream tasks. Despite modifying only 0.1\% of patches per slide with imperceptible noise, our attack leads to downstream accuracy degradation that can reach up to 20\% in the worst cases. Furthermore, we analyze key factors that influence attack success, explore the relationship between patch-level vulnerability and semantic content, and conduct a preliminary investigation into potential defence strategies. These findings lay the groundwork for future research on the adversarial robustness and reliable deployment of pathology foundation models. Our code is publicly available at: https://github.com/Jiashuai-Liu-hmos/Attack-WSI-pathology-foundation-models.
Fusing Bluetooth with Pedestrian Dead Reckoning: A Floor Plan-Assisted Positioning Approach
Apr 14, 2025
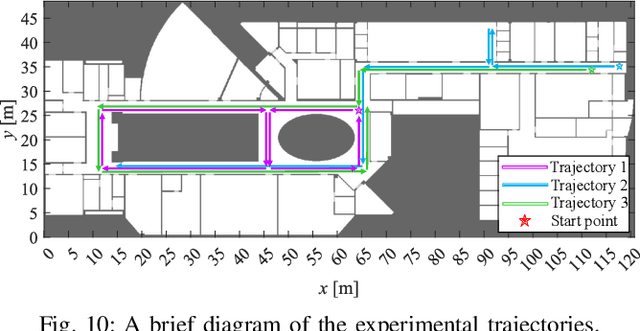
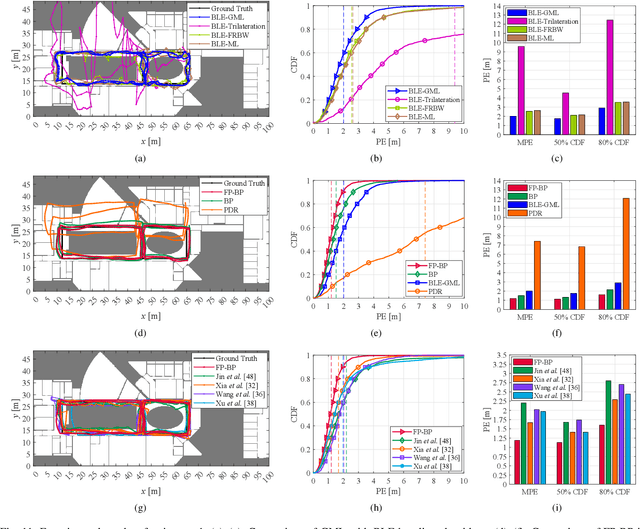
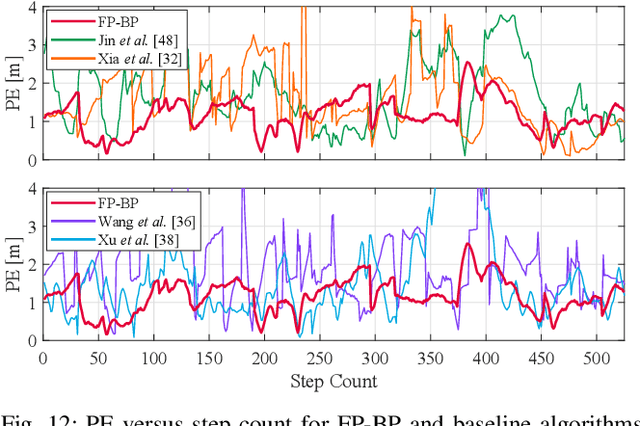
Abstract:Floor plans can provide valuable prior information that helps enhance the accuracy of indoor positioning systems. However, existing research typically faces challenges in efficiently leveraging floor plan information and applying it to complex indoor layouts. To fully exploit information from floor plans for positioning, we propose a floor plan-assisted fusion positioning algorithm (FP-BP) using Bluetooth low energy (BLE) and pedestrian dead reckoning (PDR). In the considered system, a user holding a smartphone walks through a positioning area with BLE beacons installed on the ceiling, and can locate himself in real time. In particular, FP-BP consists of two phases. In the offline phase, FP-BP programmatically extracts map features from a stylized floor plan based on their binary masks, and constructs a mapping function to identify the corresponding map feature of any given position on the map. In the online phase, FP-BP continuously computes BLE positions and PDR results from BLE signals and smartphone sensors, where a novel grid-based maximum likelihood estimation (GML) algorithm is introduced to enhance BLE positioning. Then, a particle filter is used to fuse them and obtain an initial estimate. Finally, FP-BP performs post-position correction to obtain the final position based on its specific map feature. Experimental results show that FP-BP can achieve a real-time mean positioning accuracy of 1.19 m, representing an improvement of over 28% compared to existing floor plan-fused baseline algorithms.
MapGlue: Multimodal Remote Sensing Image Matching
Mar 20, 2025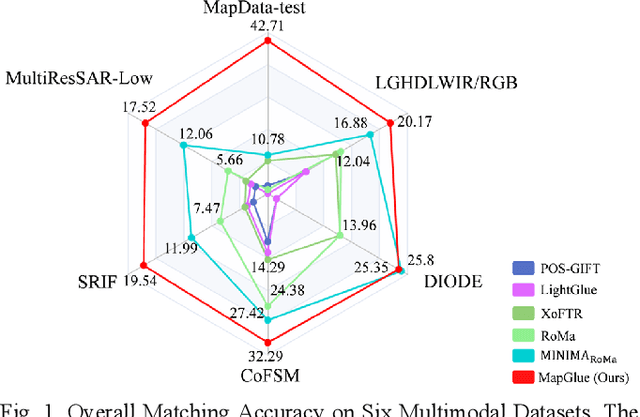
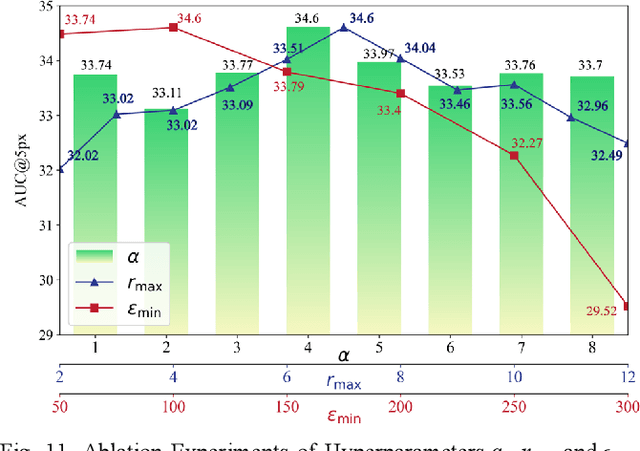
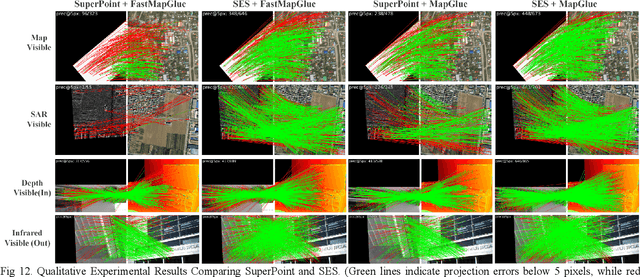
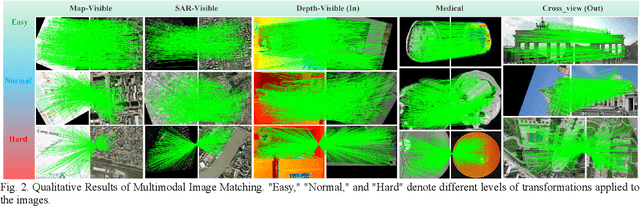
Abstract:Multimodal remote sensing image (MRSI) matching is pivotal for cross-modal fusion, localization, and object detection, but it faces severe challenges due to geometric, radiometric, and viewpoint discrepancies across imaging modalities. Existing unimodal datasets lack scale and diversity, limiting deep learning solutions. This paper proposes MapGlue, a universal MRSI matching framework, and MapData, a large-scale multimodal dataset addressing these gaps. Our contributions are twofold. MapData, a globally diverse dataset spanning 233 sampling points, offers original images (7,000x5,000 to 20,000x15,000 pixels). After rigorous cleaning, it provides 121,781 aligned electronic map-visible image pairs (512x512 pixels) with hybrid manual-automated ground truth, addressing the scarcity of scalable multimodal benchmarks. MapGlue integrates semantic context with a dual graph-guided mechanism to extract cross-modal invariant features. This structure enables global-to-local interaction, enhancing descriptor robustness against modality-specific distortions. Extensive evaluations on MapData and five public datasets demonstrate MapGlue's superiority in matching accuracy under complex conditions, outperforming state-of-the-art methods. Notably, MapGlue generalizes effectively to unseen modalities without retraining, highlighting its adaptability. This work addresses longstanding challenges in MRSI matching by combining scalable dataset construction with a robust, semantics-driven framework. Furthermore, MapGlue shows strong generalization capabilities on other modality matching tasks for which it was not specifically trained. The dataset and code are available at https://github.com/PeihaoWu/MapGlue.
Unlocking the Potential of Weakly Labeled Data: A Co-Evolutionary Learning Framework for Abnormality Detection and Report Generation
Dec 18, 2024



Abstract:Anatomical abnormality detection and report generation of chest X-ray (CXR) are two essential tasks in clinical practice. The former aims at localizing and characterizing cardiopulmonary radiological findings in CXRs, while the latter summarizes the findings in a detailed report for further diagnosis and treatment. Existing methods often focused on either task separately, ignoring their correlation. This work proposes a co-evolutionary abnormality detection and report generation (CoE-DG) framework. The framework utilizes both fully labeled (with bounding box annotations and clinical reports) and weakly labeled (with reports only) data to achieve mutual promotion between the abnormality detection and report generation tasks. Specifically, we introduce a bi-directional information interaction strategy with generator-guided information propagation (GIP) and detector-guided information propagation (DIP). For semi-supervised abnormality detection, GIP takes the informative feature extracted by the generator as an auxiliary input to the detector and uses the generator's prediction to refine the detector's pseudo labels. We further propose an intra-image-modal self-adaptive non-maximum suppression module (SA-NMS). This module dynamically rectifies pseudo detection labels generated by the teacher detection model with high-confidence predictions by the student.Inversely, for report generation, DIP takes the abnormalities' categories and locations predicted by the detector as input and guidance for the generator to improve the generated reports.
Cross-View Geo-Localization with Street-View and VHR Satellite Imagery in Decentrality Settings
Dec 16, 2024Abstract:Cross-View Geo-Localization tackles the problem of image geo-localization in GNSS-denied environments by matching street-view query images with geo-tagged aerial-view reference images. However, existing datasets and methods often assume center-aligned settings or only consider limited decentrality (i.e., the offset of the query image from the reference image center). This assumption overlooks the challenges present in real-world applications, where large decentrality can significantly enhance localization efficiency but simultaneously lead to a substantial degradation in localization accuracy. To address this limitation, we introduce CVSat, a novel dataset designed to evaluate cross-view geo-localization with a large geographic scope and diverse landscapes, emphasizing the decentrality issue. Meanwhile, we propose AuxGeo (Auxiliary Enhanced Geo-Localization), which leverages a multi-metric optimization strategy with two novel modules: the Bird's-eye view Intermediary Module (BIM) and the Position Constraint Module (PCM). BIM uses bird's-eye view images derived from street-view panoramas as an intermediary, simplifying the cross-view challenge with decentrality to a cross-view problem and a decentrality problem. PCM leverages position priors between cross-view images to establish multi-grained alignment constraints. These modules improve the performance of cross-view geo-localization with the decentrality problem. Extensive experiments demonstrate that AuxGeo outperforms previous methods on our proposed CVSat dataset, mitigating the issue of large decentrality, and also achieves state-of-the-art performance on existing public datasets such as CVUSA, CVACT, and VIGOR.
MoME: Mixture of Multimodal Experts for Cancer Survival Prediction
Jun 14, 2024


Abstract:Survival analysis, as a challenging task, requires integrating Whole Slide Images (WSIs) and genomic data for comprehensive decision-making. There are two main challenges in this task: significant heterogeneity and complex inter- and intra-modal interactions between the two modalities. Previous approaches utilize co-attention methods, which fuse features from both modalities only once after separate encoding. However, these approaches are insufficient for modeling the complex task due to the heterogeneous nature between the modalities. To address these issues, we propose a Biased Progressive Encoding (BPE) paradigm, performing encoding and fusion simultaneously. This paradigm uses one modality as a reference when encoding the other. It enables deep fusion of the modalities through multiple alternating iterations, progressively reducing the cross-modal disparities and facilitating complementary interactions. Besides modality heterogeneity, survival analysis involves various biomarkers from WSIs, genomics, and their combinations. The critical biomarkers may exist in different modalities under individual variations, necessitating flexible adaptation of the models to specific scenarios. Therefore, we further propose a Mixture of Multimodal Experts (MoME) layer to dynamically selects tailored experts in each stage of the BPE paradigm. Experts incorporate reference information from another modality to varying degrees, enabling a balanced or biased focus on different modalities during the encoding process. Extensive experimental results demonstrate the superior performance of our method on various datasets, including TCGA-BLCA, TCGA-UCEC and TCGA-LUAD. Codes are available at https://github.com/BearCleverProud/MoME.
Self-Supervised Learning for Medical Image Data with Anatomy-Oriented Imaging Planes
Apr 07, 2024



Abstract:Self-supervised learning has emerged as a powerful tool for pretraining deep networks on unlabeled data, prior to transfer learning of target tasks with limited annotation. The relevance between the pretraining pretext and target tasks is crucial to the success of transfer learning. Various pretext tasks have been proposed to utilize properties of medical image data (e.g., three dimensionality), which are more relevant to medical image analysis than generic ones for natural images. However, previous work rarely paid attention to data with anatomy-oriented imaging planes, e.g., standard cardiac magnetic resonance imaging views. As these imaging planes are defined according to the anatomy of the imaged organ, pretext tasks effectively exploiting this information can pretrain the networks to gain knowledge on the organ of interest. In this work, we propose two complementary pretext tasks for this group of medical image data based on the spatial relationship of the imaging planes. The first is to learn the relative orientation between the imaging planes and implemented as regressing their intersecting lines. The second exploits parallel imaging planes to regress their relative slice locations within a stack. Both pretext tasks are conceptually straightforward and easy to implement, and can be combined in multitask learning for better representation learning. Thorough experiments on two anatomical structures (heart and knee) and representative target tasks (semantic segmentation and classification) demonstrate that the proposed pretext tasks are effective in pretraining deep networks for remarkably boosted performance on the target tasks, and superior to other recent approaches.
Federated Modality-specific Encoders and Multimodal Anchors for Personalized Brain Tumor Segmentation
Mar 18, 2024



Abstract:Most existing federated learning (FL) methods for medical image analysis only considered intramodal heterogeneity, limiting their applicability to multimodal imaging applications. In practice, it is not uncommon that some FL participants only possess a subset of the complete imaging modalities, posing inter-modal heterogeneity as a challenge to effectively training a global model on all participants' data. In addition, each participant would expect to obtain a personalized model tailored for its local data characteristics from the FL in such a scenario. In this work, we propose a new FL framework with federated modality-specific encoders and multimodal anchors (FedMEMA) to simultaneously address the two concurrent issues. Above all, FedMEMA employs an exclusive encoder for each modality to account for the inter-modal heterogeneity in the first place. In the meantime, while the encoders are shared by the participants, the decoders are personalized to meet individual needs. Specifically, a server with full-modal data employs a fusion decoder to aggregate and fuse representations from all modality-specific encoders, thus bridging the modalities to optimize the encoders via backpropagation reversely. Meanwhile, multiple anchors are extracted from the fused multimodal representations and distributed to the clients in addition to the encoder parameters. On the other end, the clients with incomplete modalities calibrate their missing-modal representations toward the global full-modal anchors via scaled dot-product cross-attention, making up the information loss due to absent modalities while adapting the representations of present ones. FedMEMA is validated on the BraTS 2020 benchmark for multimodal brain tumor segmentation. Results show that it outperforms various up-to-date methods for multimodal and personalized FL and that its novel designs are effective. Our code is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge