Donghuan Lu
IQE-CLIP: Instance-aware Query Embedding for Zero-/Few-shot Anomaly Detection in Medical Domain
Jun 12, 2025Abstract:Recent advances in vision-language models, such as CLIP, have significantly improved performance in zero- and few-shot anomaly detection (ZFSAD) tasks. However, most existing CLIP-based methods assume prior knowledge of categories and rely on carefully designed prompts tailored to specific scenarios. While these text prompts capture semantic information in the textual space, they often fail to distinguish normal and anomalous instances in the joint embedding space. Moreover, most ZFSAD approaches focus on industrial domains, with limited exploration in medical tasks. To address these limitations, we propose IQE-CLIP, a novel framework for ZFSAD in the medical domain. We show that query embeddings integrating both textual and instance-aware visual information serve as more effective indicators of anomalies. Specifically, we introduce class-based and learnable prompting tokens to better adapt CLIP to the medical setting. Furthermore, we design an instance-aware query module that extracts region-level contextual information from both modalities, enabling the generation of anomaly-sensitive embeddings. Extensive experiments on six medical datasets demonstrate that IQE-CLIP achieves state-of-the-art performance in both zero-shot and few-shot settings. Code and data are available at \href{https://github.com/hongh0/IQE-CLIP/}{this https URL}.
Unlocking the Potential of Weakly Labeled Data: A Co-Evolutionary Learning Framework for Abnormality Detection and Report Generation
Dec 18, 2024



Abstract:Anatomical abnormality detection and report generation of chest X-ray (CXR) are two essential tasks in clinical practice. The former aims at localizing and characterizing cardiopulmonary radiological findings in CXRs, while the latter summarizes the findings in a detailed report for further diagnosis and treatment. Existing methods often focused on either task separately, ignoring their correlation. This work proposes a co-evolutionary abnormality detection and report generation (CoE-DG) framework. The framework utilizes both fully labeled (with bounding box annotations and clinical reports) and weakly labeled (with reports only) data to achieve mutual promotion between the abnormality detection and report generation tasks. Specifically, we introduce a bi-directional information interaction strategy with generator-guided information propagation (GIP) and detector-guided information propagation (DIP). For semi-supervised abnormality detection, GIP takes the informative feature extracted by the generator as an auxiliary input to the detector and uses the generator's prediction to refine the detector's pseudo labels. We further propose an intra-image-modal self-adaptive non-maximum suppression module (SA-NMS). This module dynamically rectifies pseudo detection labels generated by the teacher detection model with high-confidence predictions by the student.Inversely, for report generation, DIP takes the abnormalities' categories and locations predicted by the detector as input and guidance for the generator to improve the generated reports.
Simultaneous Alignment and Surface Regression Using Hybrid 2D-3D Networks for 3D Coherent Layer Segmentation of Retinal OCT Images with Full and Sparse Annotations
Dec 04, 2023



Abstract:Layer segmentation is important to quantitative analysis of retinal optical coherence tomography (OCT). Recently, deep learning based methods have been developed to automate this task and yield remarkable performance. However, due to the large spatial gap and potential mismatch between the B-scans of an OCT volume, all of them were based on 2D segmentation of individual B-scans, which may lose the continuity and diagnostic information of the retinal layers in 3D space. Besides, most of these methods required dense annotation of the OCT volumes, which is labor-intensive and expertise-demanding. This work presents a novel framework based on hybrid 2D-3D convolutional neural networks (CNNs) to obtain continuous 3D retinal layer surfaces from OCT volumes, which works well with both full and sparse annotations. The 2D features of individual B-scans are extracted by an encoder consisting of 2D convolutions. These 2D features are then used to produce the alignment displacement vectors and layer segmentation by two 3D decoders coupled via a spatial transformer module. Two losses are proposed to utilize the retinal layers' natural property of being smooth for B-scan alignment and layer segmentation, respectively, and are the key to the semi-supervised learning with sparse annotation. The entire framework is trained end-to-end. To the best of our knowledge, this is the first work that attempts 3D retinal layer segmentation in volumetric OCT images based on CNNs. Experiments on a synthetic dataset and three public clinical datasets show that our framework can effectively align the B-scans for potential motion correction, and achieves superior performance to state-of-the-art 2D deep learning methods in terms of both layer segmentation accuracy and cross-B-scan 3D continuity in both fully and semi-supervised settings, thus offering more clinical values than previous works.
Automatic view plane prescription for cardiac magnetic resonance imaging via supervision by spatial relationship between views
Sep 22, 2023



Abstract:Background: View planning for the acquisition of cardiac magnetic resonance (CMR) imaging remains a demanding task in clinical practice. Purpose: Existing approaches to its automation relied either on an additional volumetric image not typically acquired in clinic routine, or on laborious manual annotations of cardiac structural landmarks. This work presents a clinic-compatible, annotation-free system for automatic CMR view planning. Methods: The system mines the spatial relationship, more specifically, locates the intersecting lines, between the target planes and source views, and trains deep networks to regress heatmaps defined by distances from the intersecting lines. The intersection lines are the prescription lines prescribed by the technologists at the time of image acquisition using cardiac landmarks, and retrospectively identified from the spatial relationship. As the spatial relationship is self-contained in properly stored data, the need for additional manual annotation is eliminated. In addition, the interplay of multiple target planes predicted in a source view is utilized in a stacked hourglass architecture to gradually improve the regression. Then, a multi-view planning strategy is proposed to aggregate information from the predicted heatmaps for all the source views of a target plane, for a globally optimal prescription, mimicking the similar strategy practiced by skilled human prescribers. Results: The experiments include 181 CMR exams. Our system yields the mean angular difference and point-to-plane distance of 5.68 degrees and 3.12 mm, respectively. It not only achieves superior accuracy to existing approaches including conventional atlas-based and newer deep-learning-based in prescribing the four standard CMR planes but also demonstrates prescription of the first cardiac-anatomy-oriented plane(s) from the body-oriented scout.
You've Got Two Teachers: Co-evolutionary Image and Report Distillation for Semi-supervised Anatomical Abnormality Detection in Chest X-ray
Jul 18, 2023Abstract:Chest X-ray (CXR) anatomical abnormality detection aims at localizing and characterising cardiopulmonary radiological findings in the radiographs, which can expedite clinical workflow and reduce observational oversights. Most existing methods attempted this task in either fully supervised settings which demanded costly mass per-abnormality annotations, or weakly supervised settings which still lagged badly behind fully supervised methods in performance. In this work, we propose a co-evolutionary image and report distillation (CEIRD) framework, which approaches semi-supervised abnormality detection in CXR by grounding the visual detection results with text-classified abnormalities from paired radiology reports, and vice versa. Concretely, based on the classical teacher-student pseudo label distillation (TSD) paradigm, we additionally introduce an auxiliary report classification model, whose prediction is used for report-guided pseudo detection label refinement (RPDLR) in the primary vision detection task. Inversely, we also use the prediction of the vision detection model for abnormality-guided pseudo classification label refinement (APCLR) in the auxiliary report classification task, and propose a co-evolution strategy where the vision and report models mutually promote each other with RPDLR and APCLR performed alternatively. To this end, we effectively incorporate the weak supervision by reports into the semi-supervised TSD pipeline. Besides the cross-modal pseudo label refinement, we further propose an intra-image-modal self-adaptive non-maximum suppression, where the pseudo detection labels generated by the teacher vision model are dynamically rectified by high-confidence predictions by the student. Experimental results on the public MIMIC-CXR benchmark demonstrate CEIRD's superior performance to several up-to-date weakly and semi-supervised methods.
M3AE: Multimodal Representation Learning for Brain Tumor Segmentation with Missing Modalities
Mar 09, 2023



Abstract:Multimodal magnetic resonance imaging (MRI) provides complementary information for sub-region analysis of brain tumors. Plenty of methods have been proposed for automatic brain tumor segmentation using four common MRI modalities and achieved remarkable performance. In practice, however, it is common to have one or more modalities missing due to image corruption, artifacts, acquisition protocols, allergy to contrast agents, or simply cost. In this work, we propose a novel two-stage framework for brain tumor segmentation with missing modalities. In the first stage, a multimodal masked autoencoder (M3AE) is proposed, where both random modalities (i.e., modality dropout) and random patches of the remaining modalities are masked for a reconstruction task, for self-supervised learning of robust multimodal representations against missing modalities. To this end, we name our framework M3AE. Meanwhile, we employ model inversion to optimize a representative full-modal image at marginal extra cost, which will be used to substitute for the missing modalities and boost performance during inference. Then in the second stage, a memory-efficient self distillation is proposed to distill knowledge between heterogenous missing-modal situations while fine-tuning the model for supervised segmentation. Our M3AE belongs to the 'catch-all' genre where a single model can be applied to all possible subsets of modalities, thus is economic for both training and deployment. Extensive experiments on BraTS 2018 and 2020 datasets demonstrate its superior performance to existing state-of-the-art methods with missing modalities, as well as the efficacy of its components. Our code is available at: https://github.com/ccarliu/m3ae.
RECIST Weakly Supervised Lesion Segmentation via Label-Space Co-Training
Mar 01, 2023



Abstract:As an essential indicator for cancer progression and treatment response, tumor size is often measured following the response evaluation criteria in solid tumors (RECIST) guideline in CT slices. By marking each lesion with its longest axis and the longest perpendicular one, laborious pixel-wise manual annotation can be avoided. However, such a coarse substitute cannot provide a rich and accurate base to allow versatile quantitative analysis of lesions. To this end, we propose a novel weakly supervised framework to exploit the existing rich RECIST annotations for pixel-wise lesion segmentation. Specifically, a pair of under- and over-segmenting masks are constructed for each lesion based on its RECIST annotation and served as the label for co-training a pair of subnets, respectively, along with the proposed label-space perturbation induced consistency loss to bridge the gap between the two subnets and enable effective co-training. Extensive experiments are conducted on a public dataset to demonstrate the superiority of the proposed framework regarding the RECIST-based weakly supervised segmentation task and its universal applicability to various backbone networks.
MADAv2: Advanced Multi-Anchor Based Active Domain Adaptation Segmentation
Jan 18, 2023Abstract:Unsupervised domain adaption has been widely adopted in tasks with scarce annotated data. Unfortunately, mapping the target-domain distribution to the source-domain unconditionally may distort the essential structural information of the target-domain data, leading to inferior performance. To address this issue, we firstly propose to introduce active sample selection to assist domain adaptation regarding the semantic segmentation task. By innovatively adopting multiple anchors instead of a single centroid, both source and target domains can be better characterized as multimodal distributions, in which way more complementary and informative samples are selected from the target domain. With only a little workload to manually annotate these active samples, the distortion of the target-domain distribution can be effectively alleviated, achieving a large performance gain. In addition, a powerful semi-supervised domain adaptation strategy is proposed to alleviate the long-tail distribution problem and further improve the segmentation performance. Extensive experiments are conducted on public datasets, and the results demonstrate that the proposed approach outperforms state-of-the-art methods by large margins and achieves similar performance to the fully-supervised upperbound, i.e., 71.4% mIoU on GTA5 and 71.8% mIoU on SYNTHIA. The effectiveness of each component is also verified by thorough ablation studies.
Segmentation-guided Domain Adaptation and Data Harmonization of Multi-device Retinal Optical Coherence Tomography using Cycle-Consistent Generative Adversarial Networks
Aug 31, 2022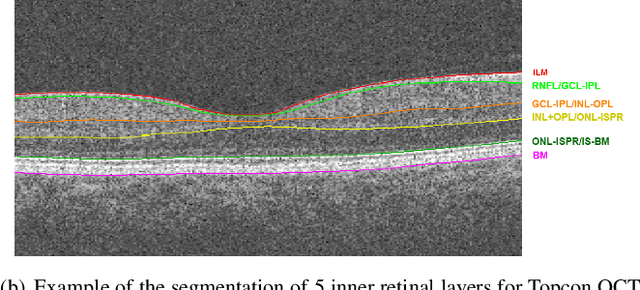
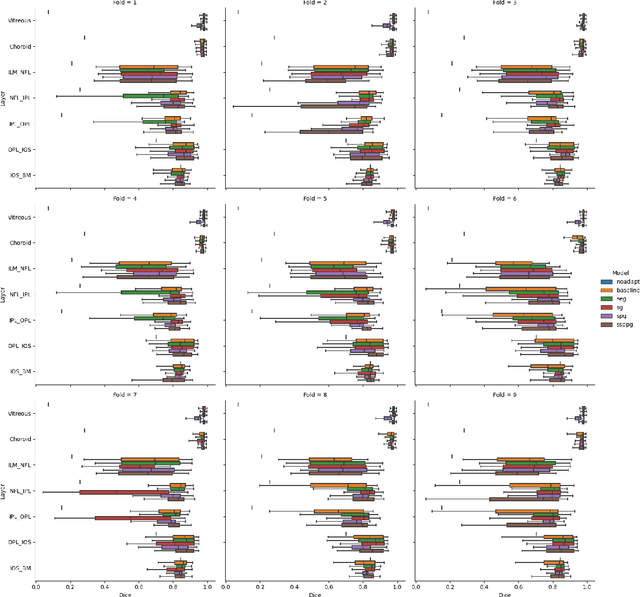
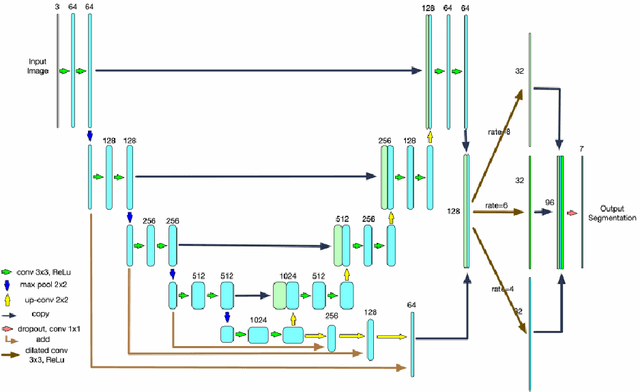
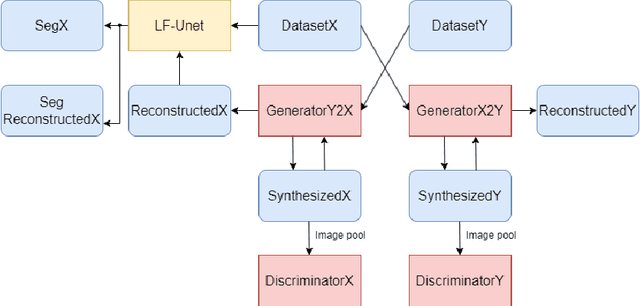
Abstract:Optical Coherence Tomography(OCT) is a non-invasive technique capturing cross-sectional area of the retina in micro-meter resolutions. It has been widely used as a auxiliary imaging reference to detect eye-related pathology and predict longitudinal progression of the disease characteristics. Retina layer segmentation is one of the crucial feature extraction techniques, where the variations of retinal layer thicknesses and the retinal layer deformation due to the presence of the fluid are highly correlated with multiple epidemic eye diseases like Diabetic Retinopathy(DR) and Age-related Macular Degeneration (AMD). However, these images are acquired from different devices, which have different intensity distribution, or in other words, belong to different imaging domains. This paper proposes a segmentation-guided domain-adaptation method to adapt images from multiple devices into single image domain, where the state-of-art pre-trained segmentation model is available. It avoids the time consumption of manual labelling for the upcoming new dataset and the re-training of the existing network. The semantic consistency and global feature consistency of the network will minimize the hallucination effect that many researchers reported regarding Cycle-Consistent Generative Adversarial Networks(CycleGAN) architecture.
Dense Cross-Query-and-Support Attention Weighted Mask Aggregation for Few-Shot Segmentation
Jul 18, 2022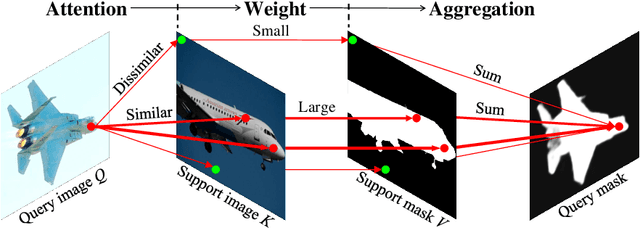


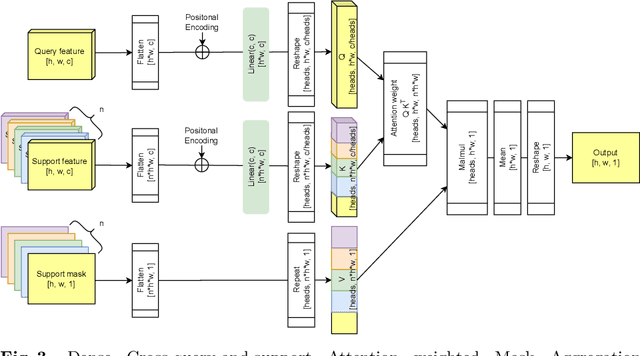
Abstract:Research into Few-shot Semantic Segmentation (FSS) has attracted great attention, with the goal to segment target objects in a query image given only a few annotated support images of the target class. A key to this challenging task is to fully utilize the information in the support images by exploiting fine-grained correlations between the query and support images. However, most existing approaches either compressed the support information into a few class-wise prototypes, or used partial support information (e.g., only foreground) at the pixel level, causing non-negligible information loss. In this paper, we propose Dense pixel-wise Cross-query-and-support Attention weighted Mask Aggregation (DCAMA), where both foreground and background support information are fully exploited via multi-level pixel-wise correlations between paired query and support features. Implemented with the scaled dot-product attention in the Transformer architecture, DCAMA treats every query pixel as a token, computes its similarities with all support pixels, and predicts its segmentation label as an additive aggregation of all the support pixels' labels -- weighted by the similarities. Based on the unique formulation of DCAMA, we further propose efficient and effective one-pass inference for n-shot segmentation, where pixels of all support images are collected for the mask aggregation at once. Experiments show that our DCAMA significantly advances the state of the art on standard FSS benchmarks of PASCAL-5i, COCO-20i, and FSS-1000, e.g., with 3.1%, 9.7%, and 3.6% absolute improvements in 1-shot mIoU over previous best records. Ablative studies also verify the design DCAMA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge