Shi Gu
Self-Supervised Learning for Medical Image Data with Anatomy-Oriented Imaging Planes
Apr 07, 2024



Abstract:Self-supervised learning has emerged as a powerful tool for pretraining deep networks on unlabeled data, prior to transfer learning of target tasks with limited annotation. The relevance between the pretraining pretext and target tasks is crucial to the success of transfer learning. Various pretext tasks have been proposed to utilize properties of medical image data (e.g., three dimensionality), which are more relevant to medical image analysis than generic ones for natural images. However, previous work rarely paid attention to data with anatomy-oriented imaging planes, e.g., standard cardiac magnetic resonance imaging views. As these imaging planes are defined according to the anatomy of the imaged organ, pretext tasks effectively exploiting this information can pretrain the networks to gain knowledge on the organ of interest. In this work, we propose two complementary pretext tasks for this group of medical image data based on the spatial relationship of the imaging planes. The first is to learn the relative orientation between the imaging planes and implemented as regressing their intersecting lines. The second exploits parallel imaging planes to regress their relative slice locations within a stack. Both pretext tasks are conceptually straightforward and easy to implement, and can be combined in multitask learning for better representation learning. Thorough experiments on two anatomical structures (heart and knee) and representative target tasks (semantic segmentation and classification) demonstrate that the proposed pretext tasks are effective in pretraining deep networks for remarkably boosted performance on the target tasks, and superior to other recent approaches.
A Concept-based Interpretable Model for the Diagnosis of Choroid Neoplasias using Multimodal Data
Mar 08, 2024Abstract:Diagnosing rare diseases presents a common challenge in clinical practice, necessitating the expertise of specialists for accurate identification. The advent of machine learning offers a promising solution, while the development of such technologies is hindered by the scarcity of data on rare conditions and the demand for models that are both interpretable and trustworthy in a clinical context. Interpretable AI, with its capacity for human-readable outputs, can facilitate validation by clinicians and contribute to medical education. In the current work, we focus on choroid neoplasias, the most prevalent form of eye cancer in adults, albeit rare with 5.1 per million. We built the so-far largest dataset consisting of 750 patients, incorporating three distinct imaging modalities collected from 2004 to 2022. Our work introduces a concept-based interpretable model that distinguishes between three types of choroidal tumors, integrating insights from domain experts via radiological reports. Remarkably, this model not only achieves an F1 score of 0.91, rivaling that of black-box models, but also boosts the diagnostic accuracy of junior doctors by 42%. This study highlights the significant potential of interpretable machine learning in improving the diagnosis of rare diseases, laying a groundwork for future breakthroughs in medical AI that could tackle a wider array of complex health scenarios.
Co-Learning Semantic-aware Unsupervised Segmentation for Pathological Image Registration
Oct 19, 2023Abstract:The registration of pathological images plays an important role in medical applications. Despite its significance, most researchers in this field primarily focus on the registration of normal tissue into normal tissue. The negative impact of focal tissue, such as the loss of spatial correspondence information and the abnormal distortion of tissue, are rarely considered. In this paper, we propose GIRNet, a novel unsupervised approach for pathological image registration by incorporating segmentation and inpainting through the principles of Generation, Inpainting, and Registration (GIR). The registration, segmentation, and inpainting modules are trained simultaneously in a co-learning manner so that the segmentation of the focal area and the registration of inpainted pairs can improve collaboratively. Overall, the registration of pathological images is achieved in a completely unsupervised learning framework. Experimental results on multiple datasets, including Magnetic Resonance Imaging (MRI) of T1 sequences, demonstrate the efficacy of our proposed method. Our results show that our method can accurately achieve the registration of pathological images and identify lesions even in challenging imaging modalities. Our unsupervised approach offers a promising solution for the efficient and cost-effective registration of pathological images. Our code is available at https://github.com/brain-intelligence-lab/GIRNet.
* 13 pages, 7 figures, published in Medical Image Computing and Computer Assisted Intervention (MICCAI) 2023
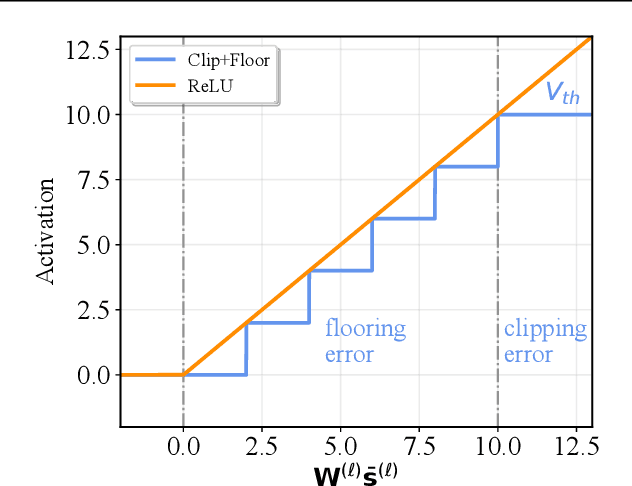
Converting Artificial Neural Networks to Spiking Neural Networks via Parameter Calibration
May 06, 2022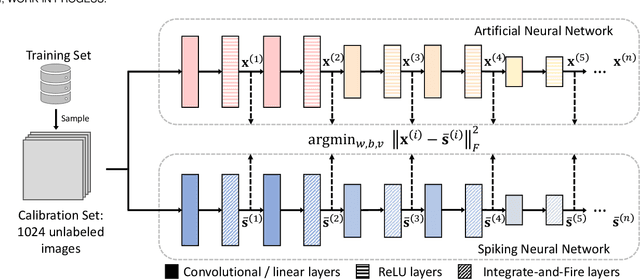
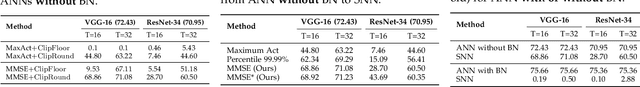
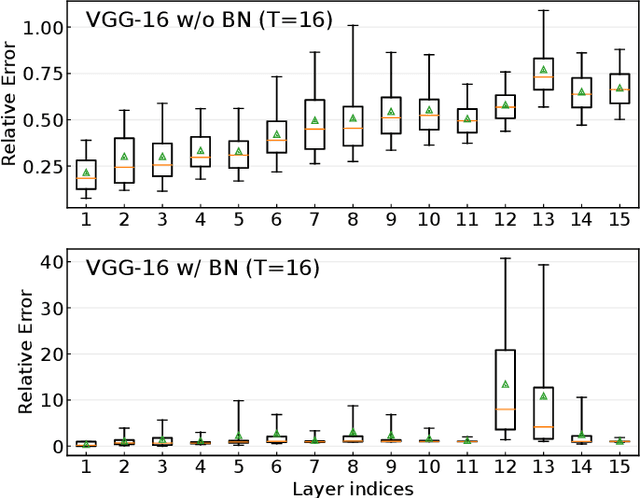
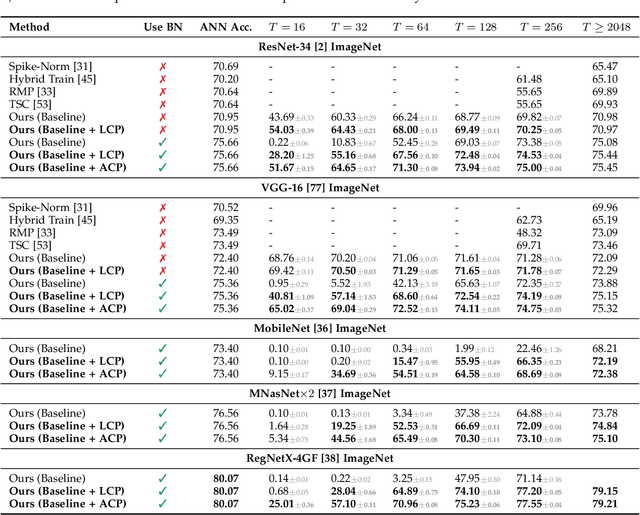
Abstract:Spiking Neural Network (SNN), originating from the neural behavior in biology, has been recognized as one of the next-generation neural networks. Conventionally, SNNs can be obtained by converting from pre-trained Artificial Neural Networks (ANNs) by replacing the non-linear activation with spiking neurons without changing the parameters. In this work, we argue that simply copying and pasting the weights of ANN to SNN inevitably results in activation mismatch, especially for ANNs that are trained with batch normalization (BN) layers. To tackle the activation mismatch issue, we first provide a theoretical analysis by decomposing local conversion error to clipping error and flooring error, and then quantitatively measure how this error propagates throughout the layers using the second-order analysis. Motivated by the theoretical results, we propose a set of layer-wise parameter calibration algorithms, which adjusts the parameters to minimize the activation mismatch. Extensive experiments for the proposed algorithms are performed on modern architectures and large-scale tasks including ImageNet classification and MS COCO detection. We demonstrate that our method can handle the SNN conversion with batch normalization layers and effectively preserve the high accuracy even in 32 time steps. For example, our calibration algorithms can increase up to 65% accuracy when converting VGG-16 with BN layers.
When Sparsity Meets Dynamic Convolution
Apr 05, 2022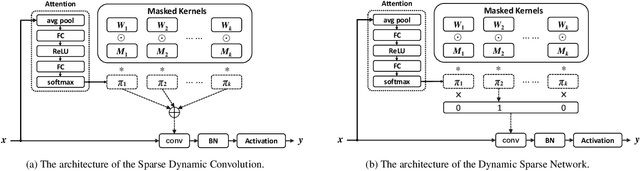
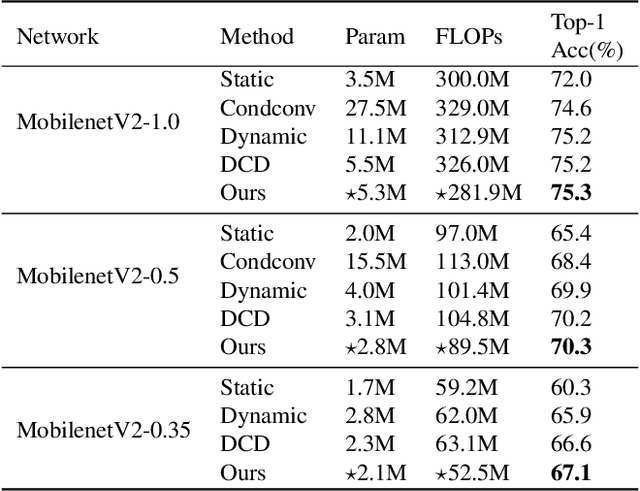
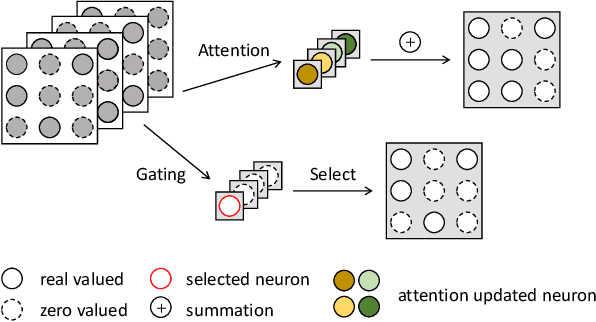

Abstract:Dynamic convolution achieves a substantial performance boost for efficient CNNs at a cost of increased convolutional weights. Contrastively, mask-based unstructured pruning obtains a lightweight network by removing redundancy in the heavy network at risk of performance drop. In this paper, we propose a new framework to coherently integrate these two paths so that they can complement each other compensate for the disadvantages. We first design a binary mask derived from a learnable threshold to prune static kernels, significantly reducing the parameters and computational cost but achieving higher performance in Imagenet-1K(0.6\% increase in top-1 accuracy with 0.67G fewer FLOPs). Based on this learnable mask, we further propose a novel dynamic sparse network incorporating the dynamic routine mechanism, which exerts much higher accuracy than baselines ($2.63\%$ increase in top-1 accuracy for MobileNetV1 with $90\%$ sparsity). As a result, our method demonstrates a more efficient dynamic convolution with sparsity.
Temporal Efficient Training of Spiking Neural Network via Gradient Re-weighting
Feb 24, 2022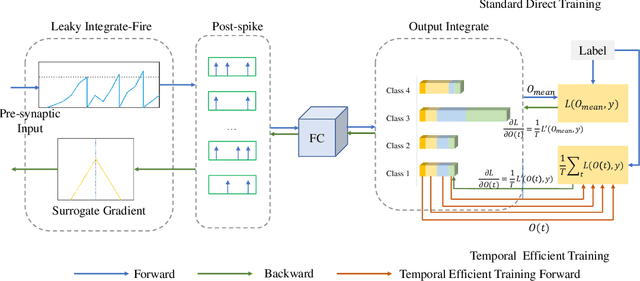
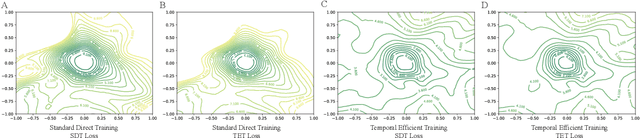
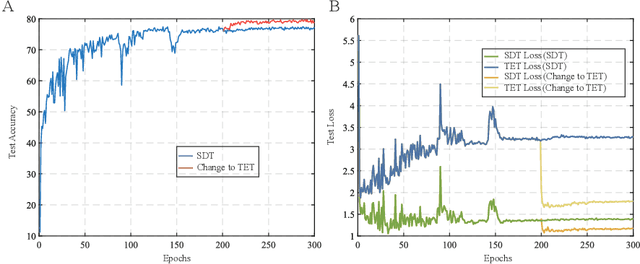
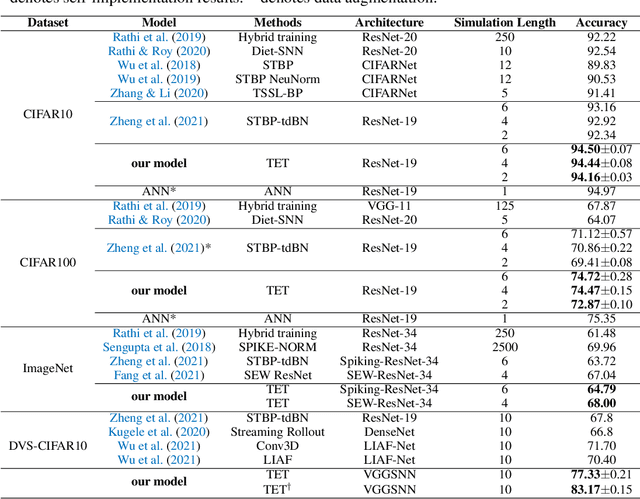
Abstract:Recently, brain-inspired spiking neuron networks (SNNs) have attracted widespread research interest because of their event-driven and energy-efficient characteristics. Still, it is difficult to efficiently train deep SNNs due to the non-differentiability of its activation function, which disables the typically used gradient descent approaches for traditional artificial neural networks (ANNs). Although the adoption of surrogate gradient (SG) formally allows for the back-propagation of losses, the discrete spiking mechanism actually differentiates the loss landscape of SNNs from that of ANNs, failing the surrogate gradient methods to achieve comparable accuracy as for ANNs. In this paper, we first analyze why the current direct training approach with surrogate gradient results in SNNs with poor generalizability. Then we introduce the temporal efficient training (TET) approach to compensate for the loss of momentum in the gradient descent with SG so that the training process can converge into flatter minima with better generalizability. Meanwhile, we demonstrate that TET improves the temporal scalability of SNN and induces a temporal inheritable training for acceleration. Our method consistently outperforms the SOTA on all reported mainstream datasets, including CIFAR-10/100 and ImageNet. Remarkably on DVS-CIFAR10, we obtained 83$\%$ top-1 accuracy, over 10$\%$ improvement compared to existing state of the art. Codes are available at \url{https://github.com/Gus-Lab/temporal_efficient_training}.
Multi-modal Attention Network for Stock Movements Prediction
Jan 14, 2022
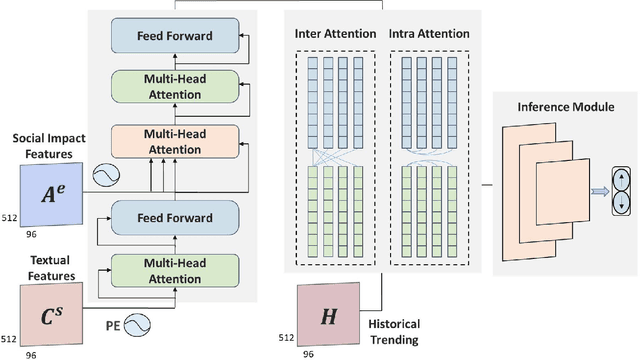
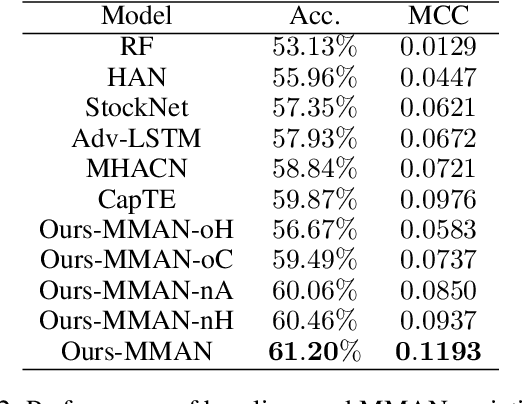
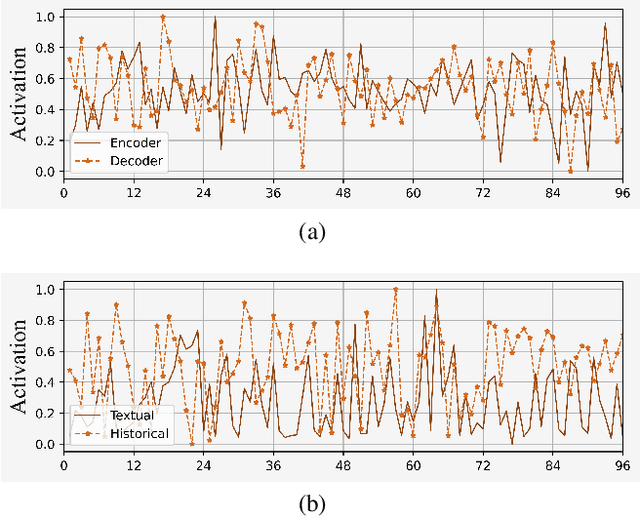
Abstract:Stock prices move as piece-wise trending fluctuation rather than a purely random walk. Traditionally, the prediction of future stock movements is based on the historical trading record. Nowadays, with the development of social media, many active participants in the market choose to publicize their strategies, which provides a window to glimpse over the whole market's attitude towards future movements by extracting the semantics behind social media. However, social media contains conflicting information and cannot replace historical records completely. In this work, we propose a multi-modality attention network to reduce conflicts and integrate semantic and numeric features to predict future stock movements comprehensively. Specifically, we first extract semantic information from social media and estimate their credibility based on posters' identity and public reputation. Then we incorporate the semantic from online posts and numeric features from historical records to make the trading strategy. Experimental results show that our approach outperforms previous methods by a significant margin in both prediction accuracy (61.20\%) and trading profits (9.13\%). It demonstrates that our method improves the performance of stock movements prediction and informs future research on multi-modality fusion towards stock prediction.
A Unified Framework for Generalized Low-Shot Medical Image Segmentation with Scarce Data
Oct 18, 2021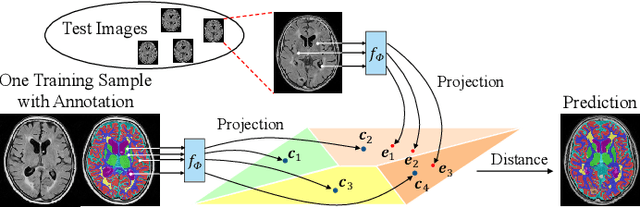
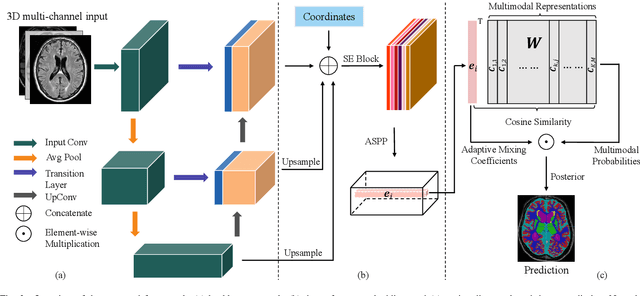
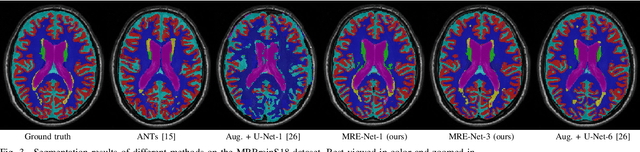
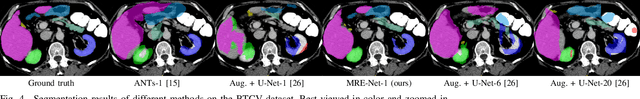
Abstract:Medical image segmentation has achieved remarkable advancements using deep neural networks (DNNs). However, DNNs often need big amounts of data and annotations for training, both of which can be difficult and costly to obtain. In this work, we propose a unified framework for generalized low-shot (one- and few-shot) medical image segmentation based on distance metric learning (DML). Unlike most existing methods which only deal with the lack of annotations while assuming abundance of data, our framework works with extreme scarcity of both, which is ideal for rare diseases. Via DML, the framework learns a multimodal mixture representation for each category, and performs dense predictions based on cosine distances between the pixels' deep embeddings and the category representations. The multimodal representations effectively utilize the inter-subject similarities and intraclass variations to overcome overfitting due to extremely limited data. In addition, we propose adaptive mixing coefficients for the multimodal mixture distributions to adaptively emphasize the modes better suited to the current input. The representations are implicitly embedded as weights of the fc layer, such that the cosine distances can be computed efficiently via forward propagation. In our experiments on brain MRI and abdominal CT datasets, the proposed framework achieves superior performances for low-shot segmentation towards standard DNN-based (3D U-Net) and classical registration-based (ANTs) methods, e.g., achieving mean Dice coefficients of 81%/69% for brain tissue/abdominal multiorgan segmentation using a single training sample, as compared to 52%/31% and 72%/35% by the U-Net and ANTs, respectively.
A Free Lunch From ANN: Towards Efficient, Accurate Spiking Neural Networks Calibration
Jun 13, 2021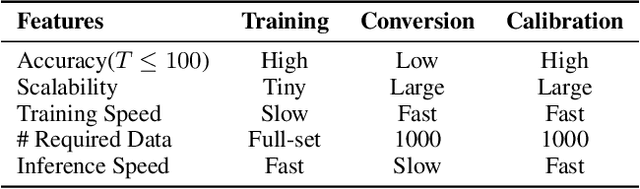
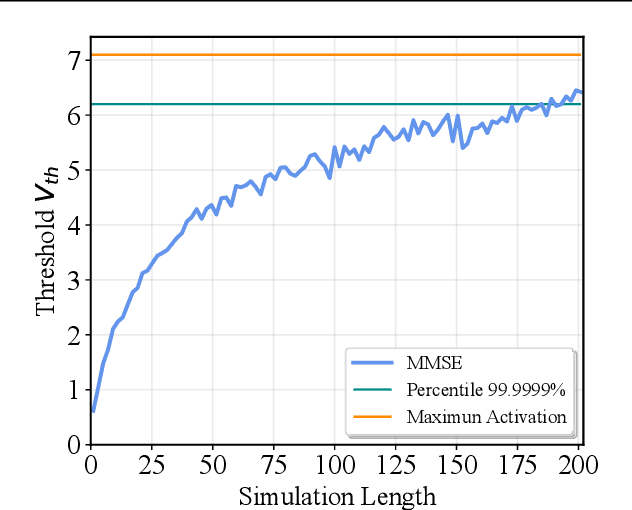
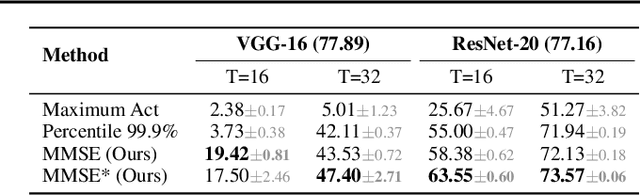
Abstract:Spiking Neural Network (SNN) has been recognized as one of the next generation of neural networks. Conventionally, SNN can be converted from a pre-trained ANN by only replacing the ReLU activation to spike activation while keeping the parameters intact. Perhaps surprisingly, in this work we show that a proper way to calibrate the parameters during the conversion of ANN to SNN can bring significant improvements. We introduce SNN Calibration, a cheap but extraordinarily effective method by leveraging the knowledge within a pre-trained Artificial Neural Network (ANN). Starting by analyzing the conversion error and its propagation through layers theoretically, we propose the calibration algorithm that can correct the error layer-by-layer. The calibration only takes a handful number of training data and several minutes to finish. Moreover, our calibration algorithm can produce SNN with state-of-the-art architecture on the large-scale ImageNet dataset, including MobileNet and RegNet. Extensive experiments demonstrate the effectiveness and efficiency of our algorithm. For example, our advanced pipeline can increase up to 69% top-1 accuracy when converting MobileNet on ImageNet compared to baselines. Codes are released at https://github.com/yhhhli/SNN_Calibration.
Optimal Conversion of Conventional Artificial Neural Networks to Spiking Neural Networks
Feb 28, 2021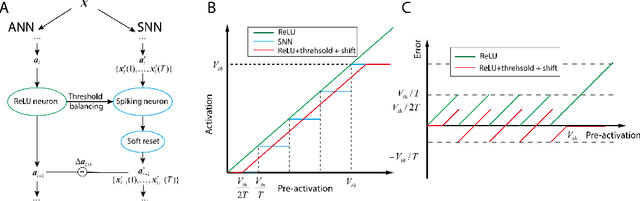
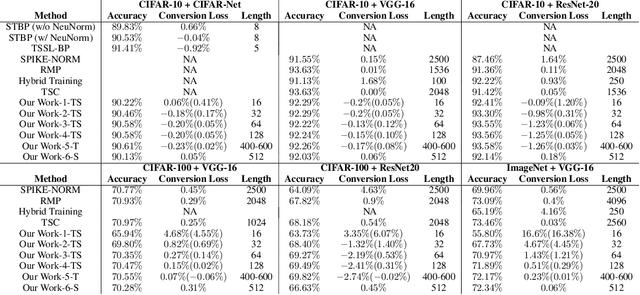
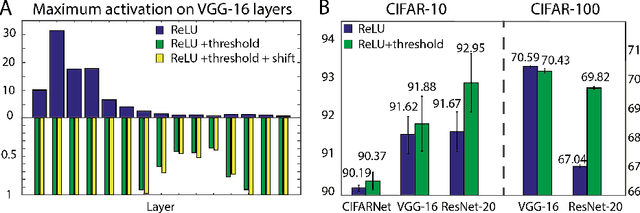
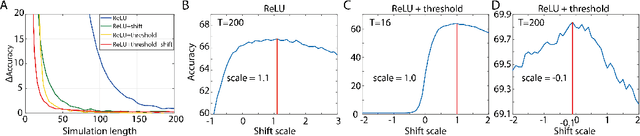
Abstract:Spiking neural networks (SNNs) are biology-inspired artificial neural networks (ANNs) that comprise of spiking neurons to process asynchronous discrete signals. While more efficient in power consumption and inference speed on the neuromorphic hardware, SNNs are usually difficult to train directly from scratch with spikes due to the discreteness. As an alternative, many efforts have been devoted to converting conventional ANNs into SNNs by copying the weights from ANNs and adjusting the spiking threshold potential of neurons in SNNs. Researchers have designed new SNN architectures and conversion algorithms to diminish the conversion error. However, an effective conversion should address the difference between the SNN and ANN architectures with an efficient approximation \DSK{of} the loss function, which is missing in the field. In this work, we analyze the conversion error by recursive reduction to layer-wise summation and propose a novel strategic pipeline that transfers the weights to the target SNN by combining threshold balance and soft-reset mechanisms. This pipeline enables almost no accuracy loss between the converted SNNs and conventional ANNs with only $\sim1/10$ of the typical SNN simulation time. Our method is promising to get implanted onto embedded platforms with better support of SNNs with limited energy and memory.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge