Conghao Xiong
ConSurv: Multimodal Continual Learning for Survival Analysis
Nov 13, 2025Abstract:Survival prediction of cancers is crucial for clinical practice, as it informs mortality risks and influences treatment plans. However, a static model trained on a single dataset fails to adapt to the dynamically evolving clinical environment and continuous data streams, limiting its practical utility. While continual learning (CL) offers a solution to learn dynamically from new datasets, existing CL methods primarily focus on unimodal inputs and suffer from severe catastrophic forgetting in survival prediction. In real-world scenarios, multimodal inputs often provide comprehensive and complementary information, such as whole slide images and genomics; and neglecting inter-modal correlations negatively impacts the performance. To address the two challenges of catastrophic forgetting and complex inter-modal interactions between gigapixel whole slide images and genomics, we propose ConSurv, the first multimodal continual learning (MMCL) method for survival analysis. ConSurv incorporates two key components: Multi-staged Mixture of Experts (MS-MoE) and Feature Constrained Replay (FCR). MS-MoE captures both task-shared and task-specific knowledge at different learning stages of the network, including two modality encoders and the modality fusion component, learning inter-modal relationships. FCR further enhances learned knowledge and mitigates forgetting by restricting feature deviation of previous data at different levels, including encoder-level features of two modalities and the fusion-level representations. Additionally, we introduce a new benchmark integrating four datasets, Multimodal Survival Analysis Incremental Learning (MSAIL), for comprehensive evaluation in the CL setting. Extensive experiments demonstrate that ConSurv outperforms competing methods across multiple metrics.
Beyond Linearity: Squeeze-and-Recalibrate Blocks for Few-Shot Whole Slide Image Classification
May 21, 2025Abstract:Deep learning has advanced computational pathology but expert annotations remain scarce. Few-shot learning mitigates annotation burdens yet suffers from overfitting and discriminative feature mischaracterization. In addition, the current few-shot multiple instance learning (MIL) approaches leverage pretrained vision-language models to alleviate these issues, but at the cost of complex preprocessing and high computational cost. We propose a Squeeze-and-Recalibrate (SR) block, a drop-in replacement for linear layers in MIL models to address these challenges. The SR block comprises two core components: a pair of low-rank trainable matrices (squeeze pathway, SP) that reduces parameter count and imposes a bottleneck to prevent spurious feature learning, and a frozen random recalibration matrix that preserves geometric structure, diversifies feature directions, and redefines the optimization objective for the SP. We provide theoretical guarantees that the SR block can approximate any linear mapping to arbitrary precision, thereby ensuring that the performance of a standard MIL model serves as a lower bound for its SR-enhanced counterpart. Extensive experiments demonstrate that our SR-MIL models consistently outperform prior methods while requiring significantly fewer parameters and no architectural changes.
A Survey of Pathology Foundation Model: Progress and Future Directions
Apr 05, 2025



Abstract:Computational pathology, analyzing whole slide images for automated cancer diagnosis, relies on the multiple instance learning framework where performance heavily depends on the feature extractor and aggregator. Recent Pathology Foundation Models (PFMs), pretrained on large-scale histopathology data, have significantly enhanced capabilities of extractors and aggregators but lack systematic analysis frameworks. This survey presents a hierarchical taxonomy organizing PFMs through a top-down philosophy that can be utilized to analyze FMs in any domain: model scope, model pretraining, and model design. Additionally, we systematically categorize PFM evaluation tasks into slide-level, patch-level, multimodal, and biological tasks, providing comprehensive benchmarking criteria. Our analysis identifies critical challenges in both PFM development (pathology-specific methodology, end-to-end pretraining, data-model scalability) and utilization (effective adaptation, model maintenance), paving the way for future directions in this promising field. Resources referenced in this survey are available at https://github.com/BearCleverProud/AwesomeWSI.
FOCUS: Knowledge-enhanced Adaptive Visual Compression for Few-shot Whole Slide Image Classification
Nov 22, 2024
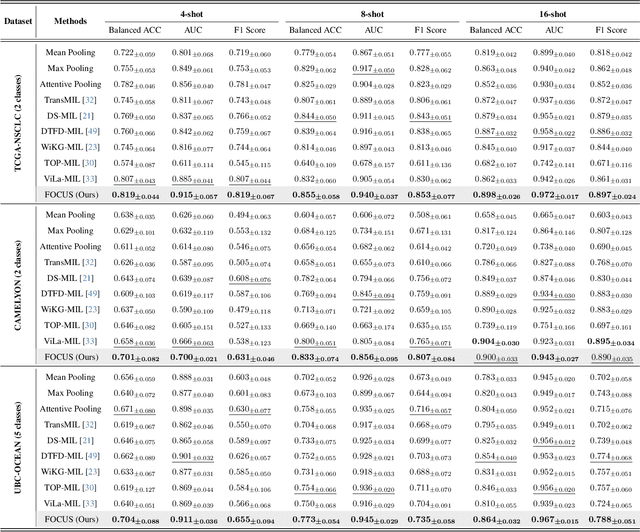
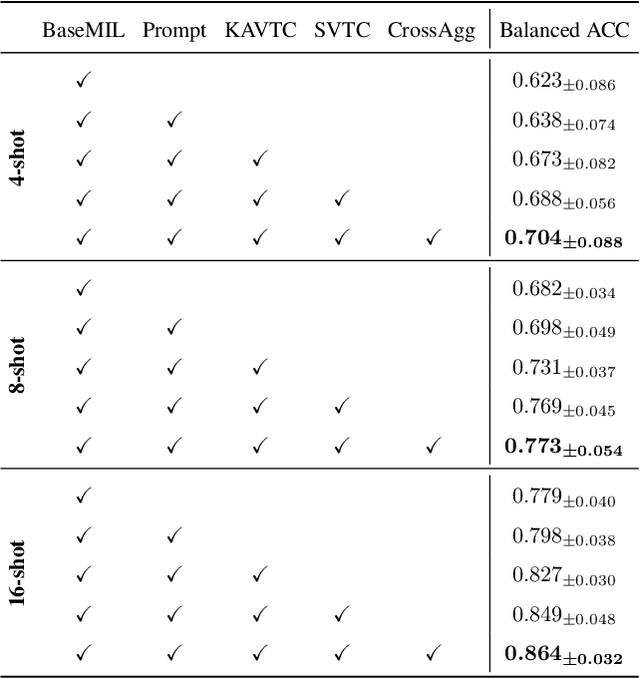
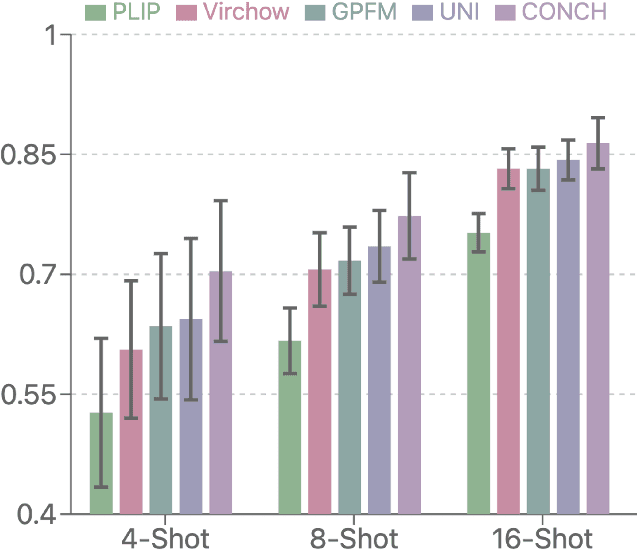
Abstract:Few-shot learning presents a critical solution for cancer diagnosis in computational pathology (CPath), addressing fundamental limitations in data availability, particularly the scarcity of expert annotations and patient privacy constraints. A key challenge in this paradigm stems from the inherent disparity between the limited training set of whole slide images (WSIs) and the enormous number of contained patches, where a significant portion of these patches lacks diagnostically relevant information, potentially diluting the model's ability to learn and focus on critical diagnostic features. While recent works attempt to address this by incorporating additional knowledge, several crucial gaps hinder further progress: (1) despite the emergence of powerful pathology foundation models (FMs), their potential remains largely untapped, with most approaches limiting their use to basic feature extraction; (2) current language guidance mechanisms attempt to align text prompts with vast numbers of WSI patches all at once, struggling to leverage rich pathological semantic information. To this end, we introduce the knowledge-enhanced adaptive visual compression framework, dubbed FOCUS, which uniquely combines pathology FMs with language prior knowledge to enable a focused analysis of diagnostically relevant regions by prioritizing discriminative WSI patches. Our approach implements a progressive three-stage compression strategy: we first leverage FMs for global visual redundancy elimination, and integrate compressed features with language prompts for semantic relevance assessment, then perform neighbor-aware visual token filtering while preserving spatial coherence. Extensive experiments on pathological datasets spanning breast, lung, and ovarian cancers demonstrate its superior performance in few-shot pathology diagnosis. Code will be made available at https://github.com/dddavid4real/FOCUS.
MoME: Mixture of Multimodal Experts for Cancer Survival Prediction
Jun 14, 2024


Abstract:Survival analysis, as a challenging task, requires integrating Whole Slide Images (WSIs) and genomic data for comprehensive decision-making. There are two main challenges in this task: significant heterogeneity and complex inter- and intra-modal interactions between the two modalities. Previous approaches utilize co-attention methods, which fuse features from both modalities only once after separate encoding. However, these approaches are insufficient for modeling the complex task due to the heterogeneous nature between the modalities. To address these issues, we propose a Biased Progressive Encoding (BPE) paradigm, performing encoding and fusion simultaneously. This paradigm uses one modality as a reference when encoding the other. It enables deep fusion of the modalities through multiple alternating iterations, progressively reducing the cross-modal disparities and facilitating complementary interactions. Besides modality heterogeneity, survival analysis involves various biomarkers from WSIs, genomics, and their combinations. The critical biomarkers may exist in different modalities under individual variations, necessitating flexible adaptation of the models to specific scenarios. Therefore, we further propose a Mixture of Multimodal Experts (MoME) layer to dynamically selects tailored experts in each stage of the BPE paradigm. Experts incorporate reference information from another modality to varying degrees, enabling a balanced or biased focus on different modalities during the encoding process. Extensive experimental results demonstrate the superior performance of our method on various datasets, including TCGA-BLCA, TCGA-UCEC and TCGA-LUAD. Codes are available at https://github.com/BearCleverProud/MoME.
Knowledge Transfer via Multi-Head Feature Adaptation for Whole Slide Image Classification
Mar 10, 2023Abstract:Transferring prior knowledge from a source domain to the same or similar target domain can greatly enhance the performance of models on the target domain. However, it is challenging to directly leverage the knowledge from the source domain due to task discrepancy and domain shift. To bridge the gaps between different tasks and domains, we propose a Multi-Head Feature Adaptation module, which projects features in the source feature space to a new space that is more similar to the target space. Knowledge transfer is particularly important in Whole Slide Image (WSI) classification since the number of WSIs in one dataset might be too small to achieve satisfactory performance. Therefore, WSI classification is an ideal testbed for our method, and we adapt multiple knowledge transfer methods for WSI classification. The experimental results show that models with knowledge transfer outperform models that are trained from scratch by a large margin regardless of the number of WSIs in the datasets, and our method achieves state-of-the-art performances among other knowledge transfer methods on multiple datasets, including TCGA-RCC, TCGA-NSCLC, and Camelyon16 datasets.
Diagnose Like a Pathologist: Transformer-Enabled Hierarchical Attention-Guided Multiple Instance Learning for Whole Slide Image Classification
Jan 19, 2023Abstract:Multiple Instance Learning (MIL) and transformers are increasingly popular in histopathology Whole Slide Image (WSI) classification. However, unlike human pathologists who selectively observe specific regions of histopathology tissues under different magnifications, most methods do not incorporate multiple resolutions of the WSIs, hierarchically and attentively, thereby leading to a loss of focus on the WSIs and information from other resolutions. To resolve this issue, we propose the Hierarchical Attention-Guided Multiple Instance Learning framework to fully exploit the WSIs, which can dynamically and attentively discover the discriminative regions across multiple resolutions of the WSIs. Within this framework, to further enhance the performance of the transformer and obtain a more holistic WSI (bag) representation, we propose an Integrated Attention Transformer, consisting of multiple Integrated Attention Modules, which is the combination of a transformer layer and an aggregation module that produces a bag representation based on every instance representation in that bag. The results of the experiments show that our method achieved state-of-the-art performances on multiple datasets, including Camelyon16, TCGA-RCC, TCGA-NSCLC, and our in-house IMGC dataset.
Embedding-Enhanced Giza++: Improving Alignment in Low- and High- Resource Scenarios Using Embedding Space Geometry
Apr 18, 2021



Abstract:A popular natural language processing task decades ago, word alignment has been dominated until recently by GIZA++, a statistical method based on the 30-year-old IBM models. Though recent years have finally seen Giza++ performance bested, the new methods primarily rely on large machine translation models, massively multilingual language models, or supervision from Giza++ alignments itself. We introduce Embedding-Enhanced Giza++, and outperform Giza++ without any of the aforementioned factors. Taking advantage of monolingual embedding space geometry of the source and target language only, we exceed Giza++'s performance in every tested scenario for three languages. In the lowest-resource scenario of only 500 lines of bitext, we improve performance over Giza++ by 10.9 AER. Our method scales monotonically outperforming Giza++ for all tested scenarios between 500 and 1.9 million lines of bitext. Our code will be made publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge