Zhengrui Guo
A Versatile Pathology Co-pilot via Reasoning Enhanced Multimodal Large Language Model
Jul 23, 2025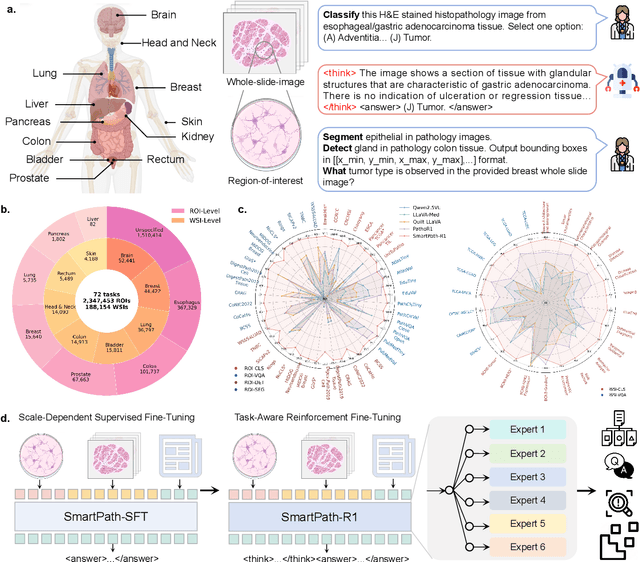
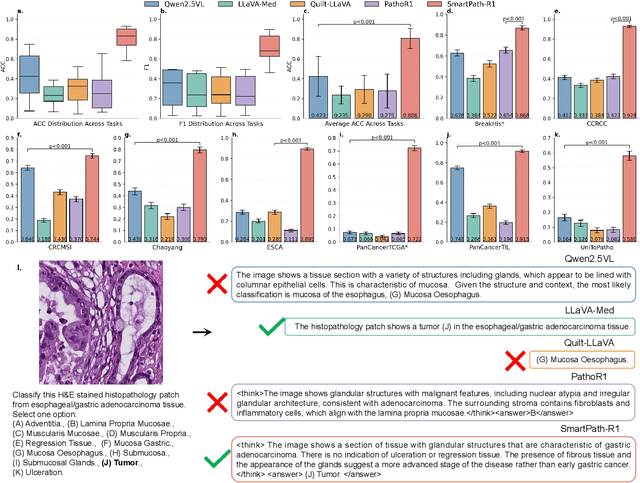
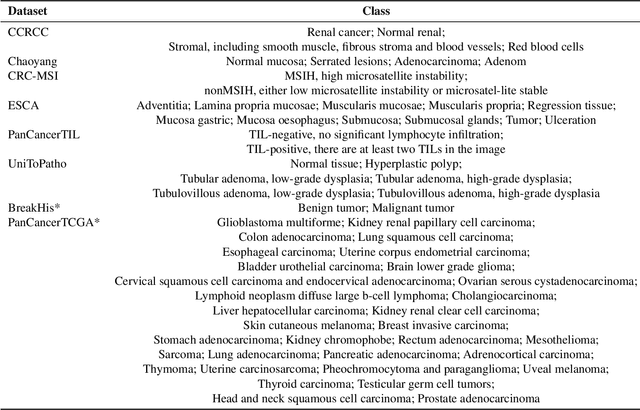
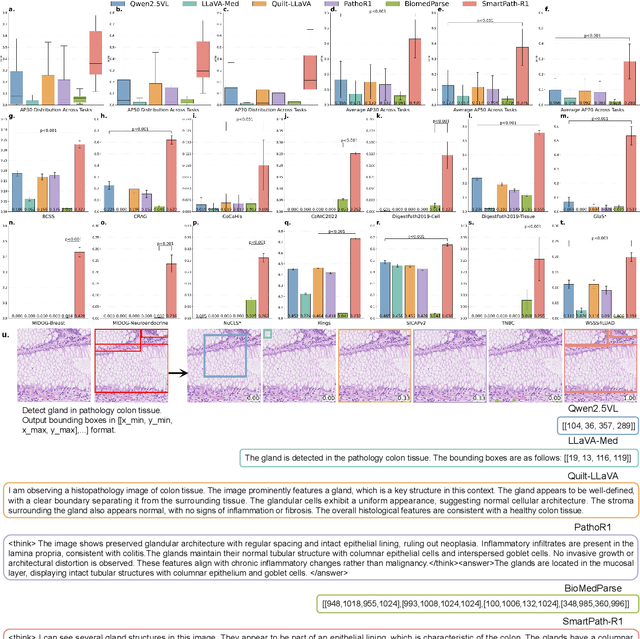
Abstract:Multimodal large language models (MLLMs) have emerged as powerful tools for computational pathology, offering unprecedented opportunities to integrate pathological images with language context for comprehensive diagnostic analysis. These models hold particular promise for automating complex tasks that traditionally require expert interpretation of pathologists. However, current MLLM approaches in pathology demonstrate significantly constrained reasoning capabilities, primarily due to their reliance on expensive chain-of-thought annotations. Additionally, existing methods remain limited to simplex application of visual question answering (VQA) at region-of-interest (ROI) level, failing to address the full spectrum of diagnostic needs such as ROI classification, detection, segmentation, whole-slide-image (WSI) classification and VQA in clinical practice. In this study, we present SmartPath-R1, a versatile MLLM capable of simultaneously addressing both ROI-level and WSI-level tasks while demonstrating robust pathological reasoning capability. Our framework combines scale-dependent supervised fine-tuning and task-aware reinforcement fine-tuning, which circumvents the requirement for chain-of-thought supervision by leveraging the intrinsic knowledge within MLLM. Furthermore, SmartPath-R1 integrates multiscale and multitask analysis through a mixture-of-experts mechanism, enabling dynamic processing for diverse tasks. We curate a large-scale dataset comprising 2.3M ROI samples and 188K WSI samples for training and evaluation. Extensive experiments across 72 tasks validate the effectiveness and superiority of the proposed approach. This work represents a significant step toward developing versatile, reasoning-enhanced AI systems for precision pathology.
PathBench: A comprehensive comparison benchmark for pathology foundation models towards precision oncology
May 26, 2025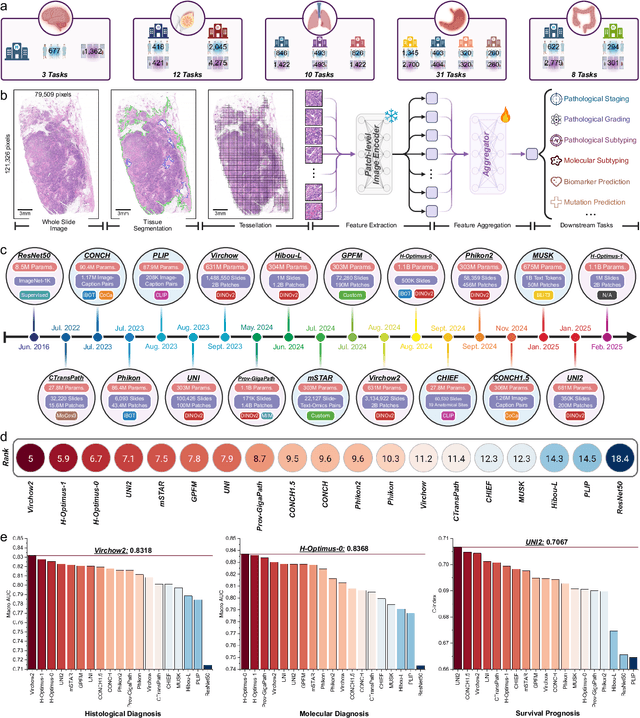



Abstract:The emergence of pathology foundation models has revolutionized computational histopathology, enabling highly accurate, generalized whole-slide image analysis for improved cancer diagnosis, and prognosis assessment. While these models show remarkable potential across cancer diagnostics and prognostics, their clinical translation faces critical challenges including variability in optimal model across cancer types, potential data leakage in evaluation, and lack of standardized benchmarks. Without rigorous, unbiased evaluation, even the most advanced PFMs risk remaining confined to research settings, delaying their life-saving applications. Existing benchmarking efforts remain limited by narrow cancer-type focus, potential pretraining data overlaps, or incomplete task coverage. We present PathBench, the first comprehensive benchmark addressing these gaps through: multi-center in-hourse datasets spanning common cancers with rigorous leakage prevention, evaluation across the full clinical spectrum from diagnosis to prognosis, and an automated leaderboard system for continuous model assessment. Our framework incorporates large-scale data, enabling objective comparison of PFMs while reflecting real-world clinical complexity. All evaluation data comes from private medical providers, with strict exclusion of any pretraining usage to avoid data leakage risks. We have collected 15,888 WSIs from 8,549 patients across 10 hospitals, encompassing over 64 diagnosis and prognosis tasks. Currently, our evaluation of 19 PFMs shows that Virchow2 and H-Optimus-1 are the most effective models overall. This work provides researchers with a robust platform for model development and offers clinicians actionable insights into PFM performance across diverse clinical scenarios, ultimately accelerating the translation of these transformative technologies into routine pathology practice.
Beyond Linearity: Squeeze-and-Recalibrate Blocks for Few-Shot Whole Slide Image Classification
May 21, 2025Abstract:Deep learning has advanced computational pathology but expert annotations remain scarce. Few-shot learning mitigates annotation burdens yet suffers from overfitting and discriminative feature mischaracterization. In addition, the current few-shot multiple instance learning (MIL) approaches leverage pretrained vision-language models to alleviate these issues, but at the cost of complex preprocessing and high computational cost. We propose a Squeeze-and-Recalibrate (SR) block, a drop-in replacement for linear layers in MIL models to address these challenges. The SR block comprises two core components: a pair of low-rank trainable matrices (squeeze pathway, SP) that reduces parameter count and imposes a bottleneck to prevent spurious feature learning, and a frozen random recalibration matrix that preserves geometric structure, diversifies feature directions, and redefines the optimization objective for the SP. We provide theoretical guarantees that the SR block can approximate any linear mapping to arbitrary precision, thereby ensuring that the performance of a standard MIL model serves as a lower bound for its SR-enhanced counterpart. Extensive experiments demonstrate that our SR-MIL models consistently outperform prior methods while requiring significantly fewer parameters and no architectural changes.
Any-to-Any Learning in Computational Pathology via Triplet Multimodal Pretraining
May 19, 2025Abstract:Recent advances in computational pathology and artificial intelligence have significantly enhanced the utilization of gigapixel whole-slide images and and additional modalities (e.g., genomics) for pathological diagnosis. Although deep learning has demonstrated strong potential in pathology, several key challenges persist: (1) fusing heterogeneous data types requires sophisticated strategies beyond simple concatenation due to high computational costs; (2) common scenarios of missing modalities necessitate flexible strategies that allow the model to learn robustly in the absence of certain modalities; (3) the downstream tasks in CPath are diverse, ranging from unimodal to multimodal, cnecessitating a unified model capable of handling all modalities. To address these challenges, we propose ALTER, an any-to-any tri-modal pretraining framework that integrates WSIs, genomics, and pathology reports. The term "any" emphasizes ALTER's modality-adaptive design, enabling flexible pretraining with any subset of modalities, and its capacity to learn robust, cross-modal representations beyond WSI-centric approaches. We evaluate ALTER across extensive clinical tasks including survival prediction, cancer subtyping, gene mutation prediction, and report generation, achieving superior or comparable performance to state-of-the-art baselines.
Context Matters: Query-aware Dynamic Long Sequence Modeling of Gigapixel Images
Jan 31, 2025Abstract:Whole slide image (WSI) analysis presents significant computational challenges due to the massive number of patches in gigapixel images. While transformer architectures excel at modeling long-range correlations through self-attention, their quadratic computational complexity makes them impractical for computational pathology applications. Existing solutions like local-global or linear self-attention reduce computational costs but compromise the strong modeling capabilities of full self-attention. In this work, we propose Querent, i.e., the query-aware long contextual dynamic modeling framework, which maintains the expressive power of full self-attention while achieving practical efficiency. Our method adaptively predicts which surrounding regions are most relevant for each patch, enabling focused yet unrestricted attention computation only with potentially important contexts. By using efficient region-wise metadata computation and importance estimation, our approach dramatically reduces computational overhead while preserving global perception to model fine-grained patch correlations. Through comprehensive experiments on biomarker prediction, gene mutation prediction, cancer subtyping, and survival analysis across over 10 WSI datasets, our method demonstrates superior performance compared to the state-of-the-art approaches. Code will be made available at https://github.com/dddavid4real/Querent.
FOCUS: Knowledge-enhanced Adaptive Visual Compression for Few-shot Whole Slide Image Classification
Nov 22, 2024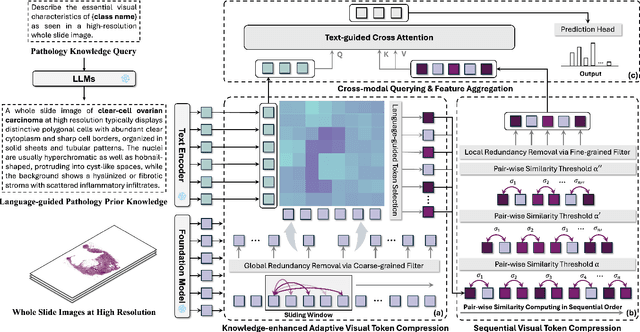
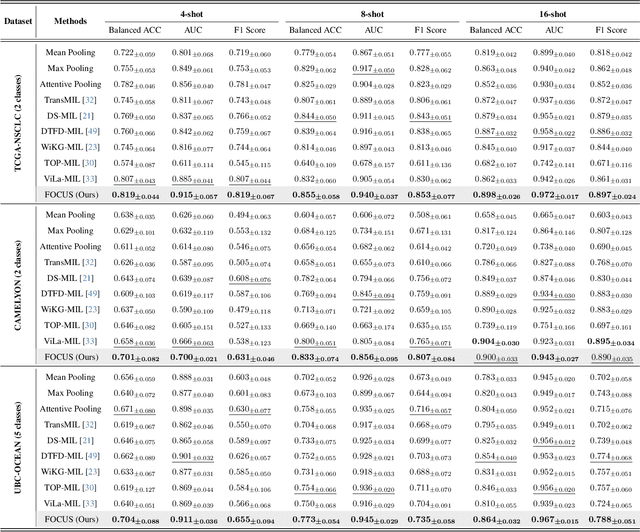
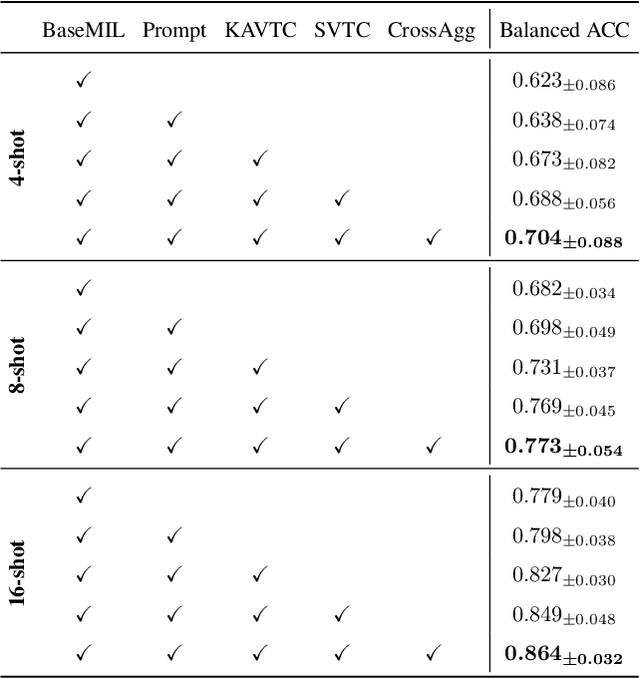
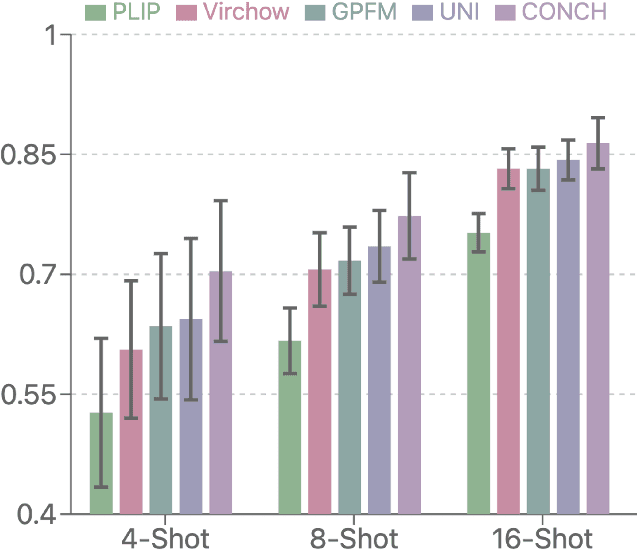
Abstract:Few-shot learning presents a critical solution for cancer diagnosis in computational pathology (CPath), addressing fundamental limitations in data availability, particularly the scarcity of expert annotations and patient privacy constraints. A key challenge in this paradigm stems from the inherent disparity between the limited training set of whole slide images (WSIs) and the enormous number of contained patches, where a significant portion of these patches lacks diagnostically relevant information, potentially diluting the model's ability to learn and focus on critical diagnostic features. While recent works attempt to address this by incorporating additional knowledge, several crucial gaps hinder further progress: (1) despite the emergence of powerful pathology foundation models (FMs), their potential remains largely untapped, with most approaches limiting their use to basic feature extraction; (2) current language guidance mechanisms attempt to align text prompts with vast numbers of WSI patches all at once, struggling to leverage rich pathological semantic information. To this end, we introduce the knowledge-enhanced adaptive visual compression framework, dubbed FOCUS, which uniquely combines pathology FMs with language prior knowledge to enable a focused analysis of diagnostically relevant regions by prioritizing discriminative WSI patches. Our approach implements a progressive three-stage compression strategy: we first leverage FMs for global visual redundancy elimination, and integrate compressed features with language prompts for semantic relevance assessment, then perform neighbor-aware visual token filtering while preserving spatial coherence. Extensive experiments on pathological datasets spanning breast, lung, and ovarian cancers demonstrate its superior performance in few-shot pathology diagnosis. Code will be made available at https://github.com/dddavid4real/FOCUS.
Towards A Generalizable Pathology Foundation Model via Unified Knowledge Distillation
Jul 26, 2024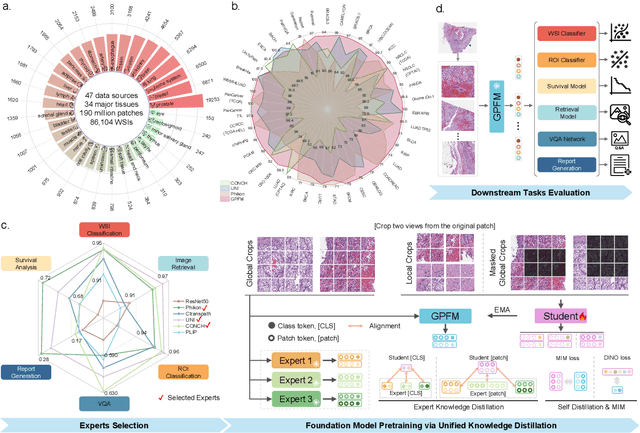

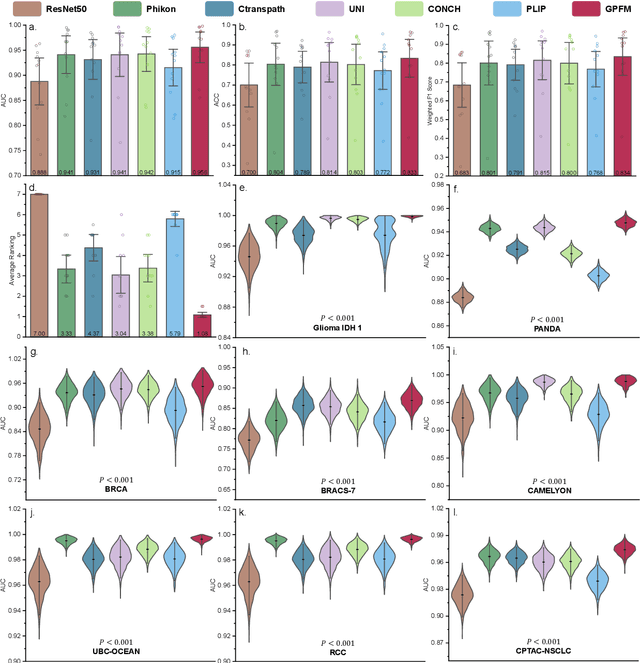
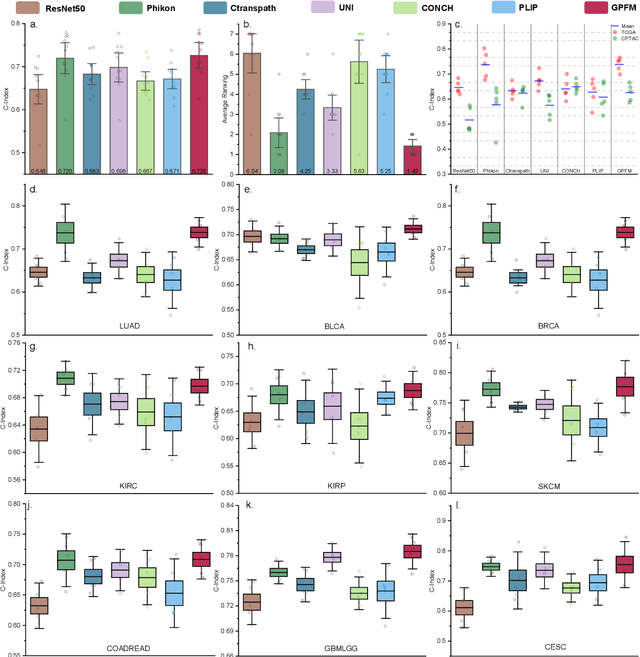
Abstract:Foundation models pretrained on large-scale datasets are revolutionizing the field of computational pathology (CPath). The generalization ability of foundation models is crucial for the success in various downstream clinical tasks. However, current foundation models have only been evaluated on a limited type and number of tasks, leaving their generalization ability and overall performance unclear. To address this gap, we established a most comprehensive benchmark to evaluate the performance of off-the-shelf foundation models across six distinct clinical task types, encompassing a total of 39 specific tasks. Our findings reveal that existing foundation models excel at certain task types but struggle to effectively handle the full breadth of clinical tasks. To improve the generalization of pathology foundation models, we propose a unified knowledge distillation framework consisting of both expert and self knowledge distillation, where the former allows the model to learn from the knowledge of multiple expert models, while the latter leverages self-distillation to enable image representation learning via local-global alignment. Based on this framework, a Generalizable Pathology Foundation Model (GPFM) is pretrained on a large-scale dataset consisting of 190 million images from around 86,000 public H\&E whole slides across 34 major tissue types. Evaluated on the established benchmark, GPFM achieves an impressive average rank of 1.36, with 29 tasks ranked 1st, while the the second-best model, UNI, attains an average rank of 2.96, with only 4 tasks ranked 1st. The superior generalization of GPFM demonstrates its exceptional modeling capabilities across a wide range of clinical tasks, positioning it as a new cornerstone for feature representation in CPath.
HistGen: Histopathology Report Generation via Local-Global Feature Encoding and Cross-modal Context Interaction
Mar 08, 2024



Abstract:Histopathology serves as the gold standard in cancer diagnosis, with clinical reports being vital in interpreting and understanding this process, guiding cancer treatment and patient care. The automation of histopathology report generation with deep learning stands to significantly enhance clinical efficiency and lessen the labor-intensive, time-consuming burden on pathologists in report writing. In pursuit of this advancement, we introduce HistGen, a multiple instance learning-empowered framework for histopathology report generation together with the first benchmark dataset for evaluation. Inspired by diagnostic and report-writing workflows, HistGen features two delicately designed modules, aiming to boost report generation by aligning whole slide images (WSIs) and diagnostic reports from local and global granularity. To achieve this, a local-global hierarchical encoder is developed for efficient visual feature aggregation from a region-to-slide perspective. Meanwhile, a cross-modal context module is proposed to explicitly facilitate alignment and interaction between distinct modalities, effectively bridging the gap between the extensive visual sequences of WSIs and corresponding highly summarized reports. Experimental results on WSI report generation show the proposed model outperforms state-of-the-art (SOTA) models by a large margin. Moreover, the results of fine-tuning our model on cancer subtyping and survival analysis tasks further demonstrate superior performance compared to SOTA methods, showcasing strong transfer learning capability. Dataset, model weights, and source code are available in https://github.com/dddavid4real/HistGen.
Label-Efficient Deep Learning in Medical Image Analysis: Challenges and Future Directions
Mar 22, 2023Abstract:Deep learning has seen rapid growth in recent years and achieved state-of-the-art performance in a wide range of applications. However, training models typically requires expensive and time-consuming collection of large quantities of labeled data. This is particularly true within the scope of medical imaging analysis (MIA), where data are limited and labels are expensive to be acquired. Thus, label-efficient deep learning methods are developed to make comprehensive use of the labeled data as well as the abundance of unlabeled and weak-labeled data. In this survey, we extensively investigated over 300 recent papers to provide a comprehensive overview of recent progress on label-efficient learning strategies in MIA. We first present the background of label-efficient learning and categorize the approaches into different schemes. Next, we examine the current state-of-the-art methods in detail through each scheme. Specifically, we provide an in-depth investigation, covering not only canonical semi-supervised, self-supervised, and multi-instance learning schemes, but also recently emerged active and annotation-efficient learning strategies. Moreover, as a comprehensive contribution to the field, this survey not only elucidates the commonalities and unique features of the surveyed methods but also presents a detailed analysis of the current challenges in the field and suggests potential avenues for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge