Fengtao Zhou
MambaMIL+: Modeling Long-Term Contextual Patterns for Gigapixel Whole Slide Image
Dec 19, 2025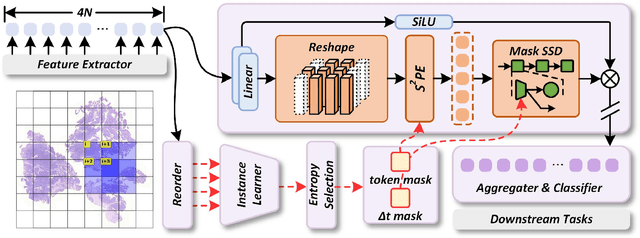
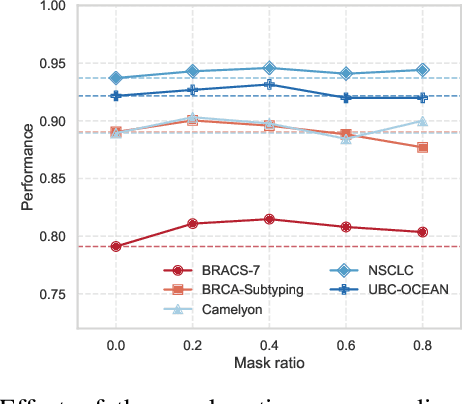
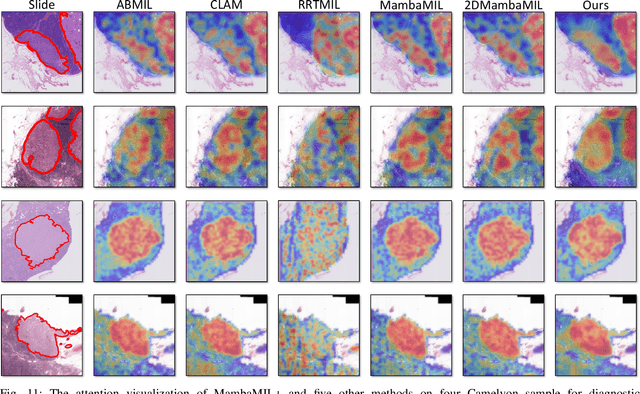
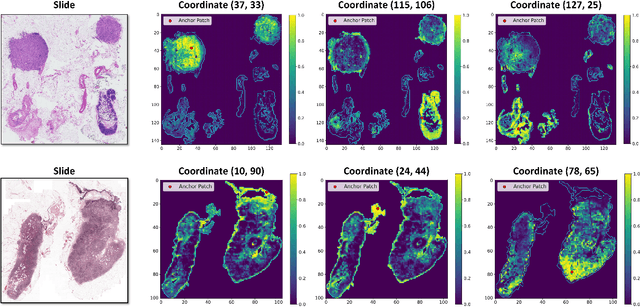
Abstract:Whole-slide images (WSIs) are an important data modality in computational pathology, yet their gigapixel resolution and lack of fine-grained annotations challenge conventional deep learning models. Multiple instance learning (MIL) offers a solution by treating each WSI as a bag of patch-level instances, but effectively modeling ultra-long sequences with rich spatial context remains difficult. Recently, Mamba has emerged as a promising alternative for long sequence learning, scaling linearly to thousands of tokens. However, despite its efficiency, it still suffers from limited spatial context modeling and memory decay, constraining its effectiveness to WSI analysis. To address these limitations, we propose MambaMIL+, a new MIL framework that explicitly integrates spatial context while maintaining long-range dependency modeling without memory forgetting. Specifically, MambaMIL+ introduces 1) overlapping scanning, which restructures the patch sequence to embed spatial continuity and instance correlations; 2) a selective stripe position encoder (S2PE) that encodes positional information while mitigating the biases of fixed scanning orders; and 3) a contextual token selection (CTS) mechanism, which leverages supervisory knowledge to dynamically enlarge the contextual memory for stable long-range modeling. Extensive experiments on 20 benchmarks across diagnostic classification, molecular prediction, and survival analysis demonstrate that MambaMIL+ consistently achieves state-of-the-art performance under three feature extractors (ResNet-50, PLIP, and CONCH), highlighting its effectiveness and robustness for large-scale computational pathology
LLM-driven Knowledge Enhancement for Multimodal Cancer Survival Prediction
Dec 16, 2025Abstract:Current multimodal survival prediction methods typically rely on pathology images (WSIs) and genomic data, both of which are high-dimensional and redundant, making it difficult to extract discriminative features from them and align different modalities. Moreover, using a simple survival follow-up label is insufficient to supervise such a complex task. To address these challenges, we propose KEMM, an LLM-driven Knowledge-Enhanced Multimodal Model for cancer survival prediction, which integrates expert reports and prognostic background knowledge. 1) Expert reports, provided by pathologists on a case-by-case basis and refined by large language model (LLM), offer succinct and clinically focused diagnostic statements. This information may typically suggest different survival outcomes. 2) Prognostic background knowledge (PBK), generated concisely by LLM, provides valuable prognostic background knowledge on different cancer types, which also enhances survival prediction. To leverage these knowledge, we introduce the knowledge-enhanced cross-modal (KECM) attention module. KECM can effectively guide the network to focus on discriminative and survival-relevant features from highly redundant modalities. Extensive experiments on five datasets demonstrate that KEMM achieves state-of-the-art performance. The code will be released upon acceptance.
A Clinical-grade Universal Foundation Model for Intraoperative Pathology
Oct 06, 2025Abstract:Intraoperative pathology is pivotal to precision surgery, yet its clinical impact is constrained by diagnostic complexity and the limited availability of high-quality frozen-section data. While computational pathology has made significant strides, the lack of large-scale, prospective validation has impeded its routine adoption in surgical workflows. Here, we introduce CRISP, a clinical-grade foundation model developed on over 100,000 frozen sections from eight medical centers, specifically designed to provide Clinical-grade Robust Intraoperative Support for Pathology (CRISP). CRISP was comprehensively evaluated on more than 15,000 intraoperative slides across nearly 100 retrospective diagnostic tasks, including benign-malignant discrimination, key intraoperative decision-making, and pan-cancer detection, etc. The model demonstrated robust generalization across diverse institutions, tumor types, and anatomical sites-including previously unseen sites and rare cancers. In a prospective cohort of over 2,000 patients, CRISP sustained high diagnostic accuracy under real-world conditions, directly informing surgical decisions in 92.6% of cases. Human-AI collaboration further reduced diagnostic workload by 35%, avoided 105 ancillary tests and enhanced detection of micrometastases with 87.5% accuracy. Together, these findings position CRISP as a clinical-grade paradigm for AI-driven intraoperative pathology, bridging computational advances with surgical precision and accelerating the translation of artificial intelligence into routine clinical practice.
Generative AI for Misalignment-Resistant Virtual Staining to Accelerate Histopathology Workflows
Sep 17, 2025Abstract:Accurate histopathological diagnosis often requires multiple differently stained tissue sections, a process that is time-consuming, labor-intensive, and environmentally taxing due to the use of multiple chemical stains. Recently, virtual staining has emerged as a promising alternative that is faster, tissue-conserving, and environmentally friendly. However, existing virtual staining methods face significant challenges in clinical applications, primarily due to their reliance on well-aligned paired data. Obtaining such data is inherently difficult because chemical staining processes can distort tissue structures, and a single tissue section cannot undergo multiple staining procedures without damage or loss of information. As a result, most available virtual staining datasets are either unpaired or roughly paired, making it difficult for existing methods to achieve accurate pixel-level supervision. To address this challenge, we propose a robust virtual staining framework featuring cascaded registration mechanisms to resolve spatial mismatches between generated outputs and their corresponding ground truth. Experimental results demonstrate that our method significantly outperforms state-of-the-art models across five datasets, achieving an average improvement of 3.2% on internal datasets and 10.1% on external datasets. Moreover, in datasets with substantial misalignment, our approach achieves a remarkable 23.8% improvement in peak signal-to-noise ratio compared to baseline models. The exceptional robustness of the proposed method across diverse datasets simplifies the data acquisition process for virtual staining and offers new insights for advancing its development.
A Multimodal Foundation Model to Enhance Generalizability and Data Efficiency for Pan-cancer Prognosis Prediction
Sep 16, 2025Abstract:Multimodal data provides heterogeneous information for a holistic understanding of the tumor microenvironment. However, existing AI models often struggle to harness the rich information within multimodal data and extract poorly generalizable representations. Here we present MICE (Multimodal data Integration via Collaborative Experts), a multimodal foundation model that effectively integrates pathology images, clinical reports, and genomics data for precise pan-cancer prognosis prediction. Instead of conventional multi-expert modules, MICE employs multiple functionally diverse experts to comprehensively capture both cross-cancer and cancer-specific insights. Leveraging data from 11,799 patients across 30 cancer types, we enhanced MICE's generalizability by coupling contrastive and supervised learning. MICE outperformed both unimodal and state-of-the-art multi-expert-based multimodal models, demonstrating substantial improvements in C-index ranging from 3.8% to 11.2% on internal cohorts and 5.8% to 8.8% on independent cohorts, respectively. Moreover, it exhibited remarkable data efficiency across diverse clinical scenarios. With its enhanced generalizability and data efficiency, MICE establishes an effective and scalable foundation for pan-cancer prognosis prediction, holding strong potential to personalize tailored therapies and improve treatment outcomes.
A Versatile Pathology Co-pilot via Reasoning Enhanced Multimodal Large Language Model
Jul 23, 2025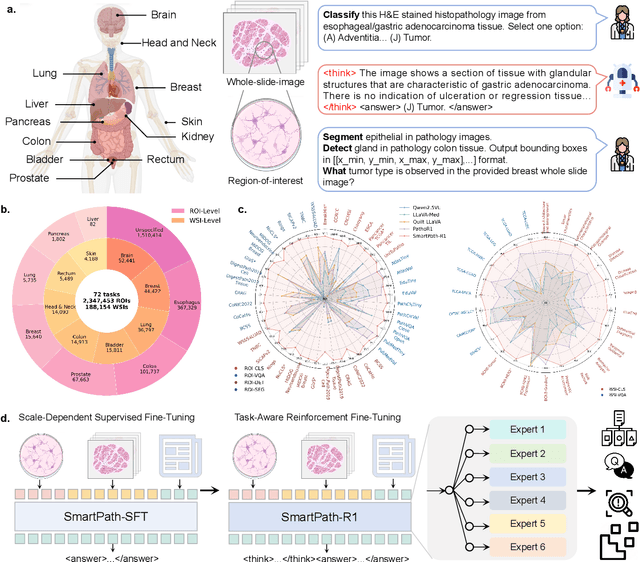
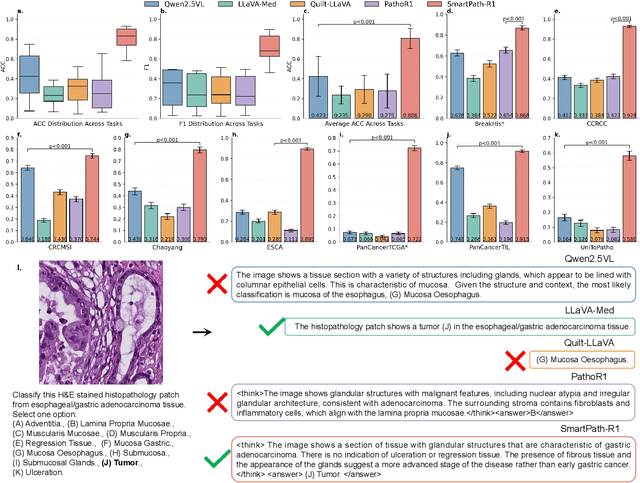
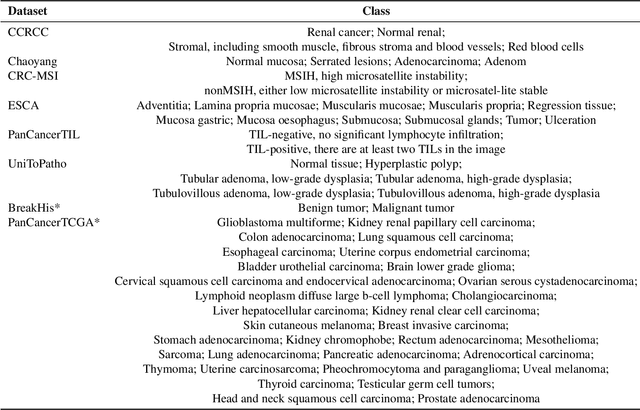
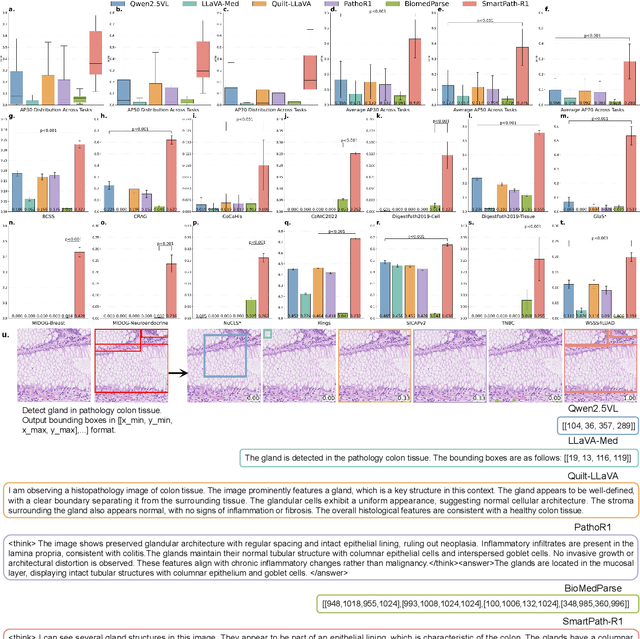
Abstract:Multimodal large language models (MLLMs) have emerged as powerful tools for computational pathology, offering unprecedented opportunities to integrate pathological images with language context for comprehensive diagnostic analysis. These models hold particular promise for automating complex tasks that traditionally require expert interpretation of pathologists. However, current MLLM approaches in pathology demonstrate significantly constrained reasoning capabilities, primarily due to their reliance on expensive chain-of-thought annotations. Additionally, existing methods remain limited to simplex application of visual question answering (VQA) at region-of-interest (ROI) level, failing to address the full spectrum of diagnostic needs such as ROI classification, detection, segmentation, whole-slide-image (WSI) classification and VQA in clinical practice. In this study, we present SmartPath-R1, a versatile MLLM capable of simultaneously addressing both ROI-level and WSI-level tasks while demonstrating robust pathological reasoning capability. Our framework combines scale-dependent supervised fine-tuning and task-aware reinforcement fine-tuning, which circumvents the requirement for chain-of-thought supervision by leveraging the intrinsic knowledge within MLLM. Furthermore, SmartPath-R1 integrates multiscale and multitask analysis through a mixture-of-experts mechanism, enabling dynamic processing for diverse tasks. We curate a large-scale dataset comprising 2.3M ROI samples and 188K WSI samples for training and evaluation. Extensive experiments across 72 tasks validate the effectiveness and superiority of the proposed approach. This work represents a significant step toward developing versatile, reasoning-enhanced AI systems for precision pathology.
Genome-Anchored Foundation Model Embeddings Improve Molecular Prediction from Histology Images
Jun 24, 2025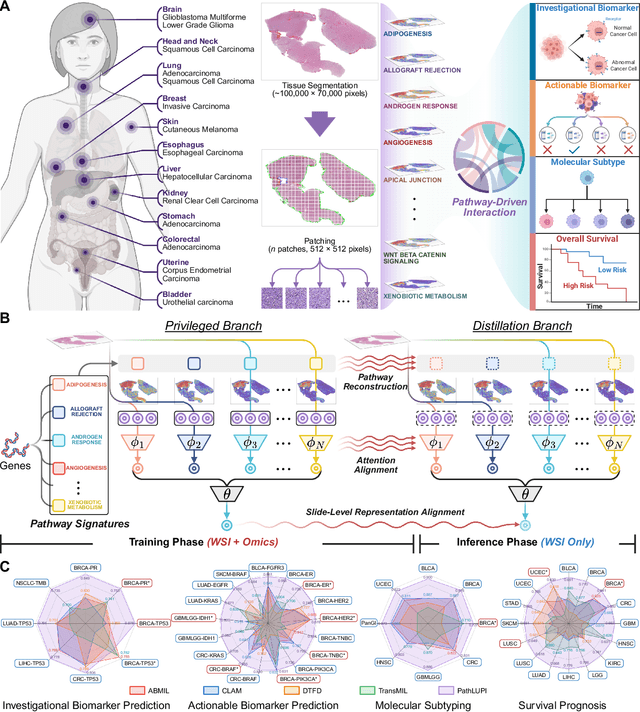
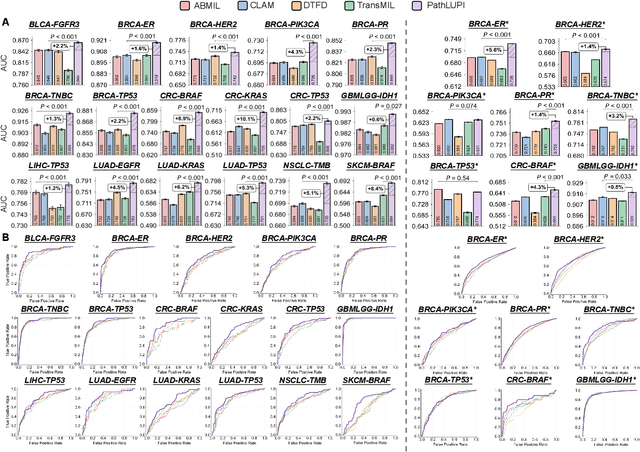
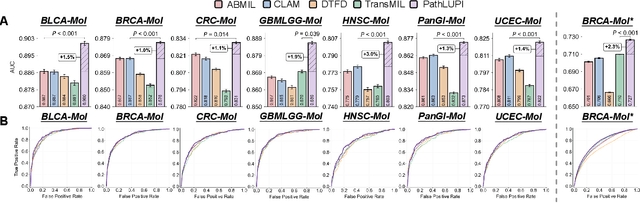
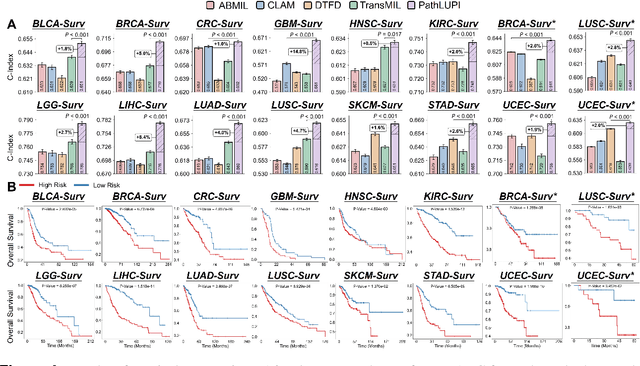
Abstract:Precision oncology requires accurate molecular insights, yet obtaining these directly from genomics is costly and time-consuming for broad clinical use. Predicting complex molecular features and patient prognosis directly from routine whole-slide images (WSI) remains a major challenge for current deep learning methods. Here we introduce PathLUPI, which uses transcriptomic privileged information during training to extract genome-anchored histological embeddings, enabling effective molecular prediction using only WSIs at inference. Through extensive evaluation across 49 molecular oncology tasks using 11,257 cases among 20 cohorts, PathLUPI demonstrated superior performance compared to conventional methods trained solely on WSIs. Crucially, it achieves AUC $\geq$ 0.80 in 14 of the biomarker prediction and molecular subtyping tasks and C-index $\geq$ 0.70 in survival cohorts of 5 major cancer types. Moreover, PathLUPI embeddings reveal distinct cellular morphological signatures associated with specific genotypes and related biological pathways within WSIs. By effectively encoding molecular context to refine WSI representations, PathLUPI overcomes a key limitation of existing models and offers a novel strategy to bridge molecular insights with routine pathology workflows for wider clinical application.
PathBench: A comprehensive comparison benchmark for pathology foundation models towards precision oncology
May 26, 2025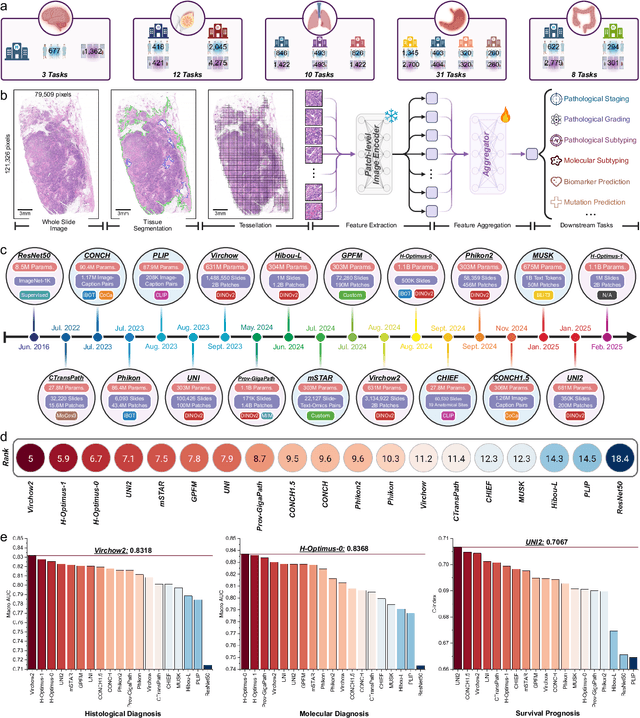



Abstract:The emergence of pathology foundation models has revolutionized computational histopathology, enabling highly accurate, generalized whole-slide image analysis for improved cancer diagnosis, and prognosis assessment. While these models show remarkable potential across cancer diagnostics and prognostics, their clinical translation faces critical challenges including variability in optimal model across cancer types, potential data leakage in evaluation, and lack of standardized benchmarks. Without rigorous, unbiased evaluation, even the most advanced PFMs risk remaining confined to research settings, delaying their life-saving applications. Existing benchmarking efforts remain limited by narrow cancer-type focus, potential pretraining data overlaps, or incomplete task coverage. We present PathBench, the first comprehensive benchmark addressing these gaps through: multi-center in-hourse datasets spanning common cancers with rigorous leakage prevention, evaluation across the full clinical spectrum from diagnosis to prognosis, and an automated leaderboard system for continuous model assessment. Our framework incorporates large-scale data, enabling objective comparison of PFMs while reflecting real-world clinical complexity. All evaluation data comes from private medical providers, with strict exclusion of any pretraining usage to avoid data leakage risks. We have collected 15,888 WSIs from 8,549 patients across 10 hospitals, encompassing over 64 diagnosis and prognosis tasks. Currently, our evaluation of 19 PFMs shows that Virchow2 and H-Optimus-1 are the most effective models overall. This work provides researchers with a robust platform for model development and offers clinicians actionable insights into PFM performance across diverse clinical scenarios, ultimately accelerating the translation of these transformative technologies into routine pathology practice.
Distilled Prompt Learning for Incomplete Multimodal Survival Prediction
Mar 03, 2025Abstract:The integration of multimodal data including pathology images and gene profiles is widely applied in precise survival prediction. Despite recent advances in multimodal survival models, collecting complete modalities for multimodal fusion still poses a significant challenge, hindering their application in clinical settings. Current approaches tackling incomplete modalities often fall short, as they typically compensate for only a limited part of the knowledge of missing modalities. To address this issue, we propose a Distilled Prompt Learning framework (DisPro) to utilize the strong robustness of Large Language Models (LLMs) to missing modalities, which employs two-stage prompting for compensation of comprehensive information for missing modalities. In the first stage, Unimodal Prompting (UniPro) distills the knowledge distribution of each modality, preparing for supplementing modality-specific knowledge of the missing modality in the subsequent stage. In the second stage, Multimodal Prompting (MultiPro) leverages available modalities as prompts for LLMs to infer the missing modality, which provides modality-common information. Simultaneously, the unimodal knowledge acquired in the first stage is injected into multimodal inference to compensate for the modality-specific knowledge of the missing modality. Extensive experiments covering various missing scenarios demonstrated the superiority of the proposed method. The code is available at https://github.com/Innse/DisPro.
Explain via Any Concept: Concept Bottleneck Model with Open Vocabulary Concepts
Aug 05, 2024



Abstract:The concept bottleneck model (CBM) is an interpretable-by-design framework that makes decisions by first predicting a set of interpretable concepts, and then predicting the class label based on the given concepts. Existing CBMs are trained with a fixed set of concepts (concepts are either annotated by the dataset or queried from language models). However, this closed-world assumption is unrealistic in practice, as users may wonder about the role of any desired concept in decision-making after the model is deployed. Inspired by the large success of recent vision-language pre-trained models such as CLIP in zero-shot classification, we propose "OpenCBM" to equip the CBM with open vocabulary concepts via: (1) Aligning the feature space of a trainable image feature extractor with that of a CLIP's image encoder via a prototype based feature alignment; (2) Simultaneously training an image classifier on the downstream dataset; (3) Reconstructing the trained classification head via any set of user-desired textual concepts encoded by CLIP's text encoder. To reveal potentially missing concepts from users, we further propose to iteratively find the closest concept embedding to the residual parameters during the reconstruction until the residual is small enough. To the best of our knowledge, our "OpenCBM" is the first CBM with concepts of open vocabularies, providing users the unique benefit such as removing, adding, or replacing any desired concept to explain the model's prediction even after a model is trained. Moreover, our model significantly outperforms the previous state-of-the-art CBM by 9% in the classification accuracy on the benchmark dataset CUB-200-2011.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge