Jiguang Wang
Fusion of Heterogeneous Pathology Foundation Models for Whole Slide Image Analysis
Oct 31, 2025Abstract:Whole slide image (WSI) analysis has emerged as an increasingly essential technique in computational pathology. Recent advances in the pathological foundation models (FMs) have demonstrated significant advantages in deriving meaningful patch-level or slide-level feature representations from WSIs. However, current pathological FMs have exhibited substantial heterogeneity caused by diverse private training datasets and different network architectures. This heterogeneity introduces performance variability when we utilize the extracted features from different FMs in the downstream tasks. To fully explore the advantage of multiple FMs effectively, in this work, we propose a novel framework for the fusion of heterogeneous pathological FMs, called FuseCPath, yielding a model with a superior ensemble performance. The main contributions of our framework can be summarized as follows: (i) To guarantee the representativeness of the training patches, we propose a multi-view clustering-based method to filter out the discriminative patches via multiple FMs' embeddings. (ii) To effectively fuse the heterogeneous patch-level FMs, we devise a cluster-level re-embedding strategy to online capture patch-level local features. (iii) To effectively fuse the heterogeneous slide-level FMs, we devise a collaborative distillation strategy to explore the connections between slide-level FMs. Extensive experiments conducted on lung cancer, bladder cancer, and colorectal cancer datasets from The Cancer Genome Atlas (TCGA) have demonstrated that the proposed FuseCPath achieves state-of-the-art performance across multiple tasks on these public datasets.
Towards A Generalizable Pathology Foundation Model via Unified Knowledge Distillation
Jul 26, 2024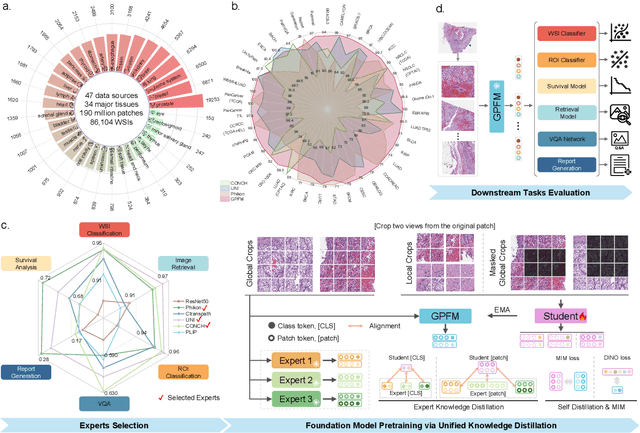

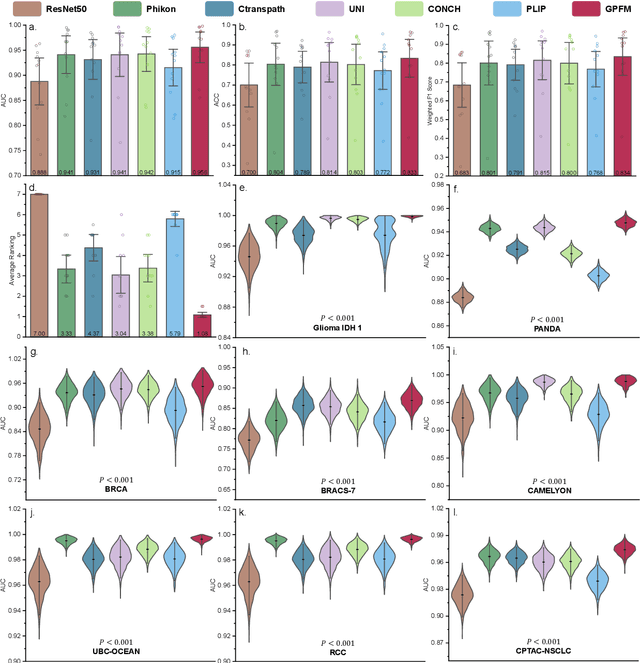
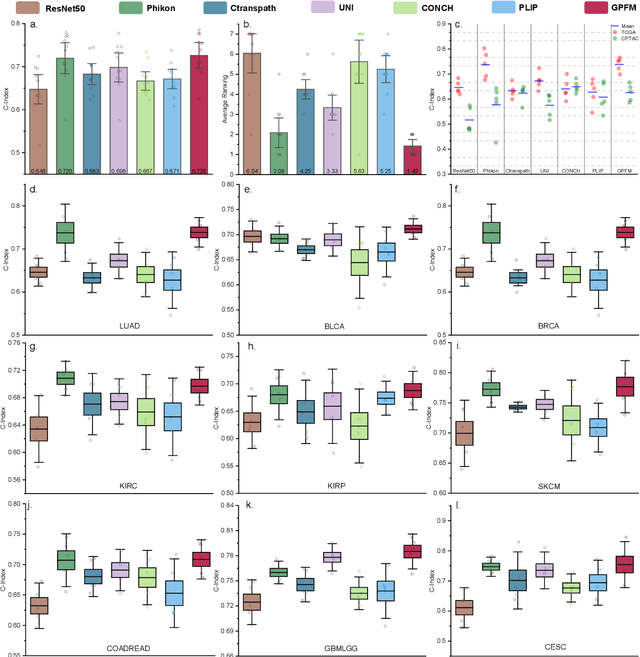
Abstract:Foundation models pretrained on large-scale datasets are revolutionizing the field of computational pathology (CPath). The generalization ability of foundation models is crucial for the success in various downstream clinical tasks. However, current foundation models have only been evaluated on a limited type and number of tasks, leaving their generalization ability and overall performance unclear. To address this gap, we established a most comprehensive benchmark to evaluate the performance of off-the-shelf foundation models across six distinct clinical task types, encompassing a total of 39 specific tasks. Our findings reveal that existing foundation models excel at certain task types but struggle to effectively handle the full breadth of clinical tasks. To improve the generalization of pathology foundation models, we propose a unified knowledge distillation framework consisting of both expert and self knowledge distillation, where the former allows the model to learn from the knowledge of multiple expert models, while the latter leverages self-distillation to enable image representation learning via local-global alignment. Based on this framework, a Generalizable Pathology Foundation Model (GPFM) is pretrained on a large-scale dataset consisting of 190 million images from around 86,000 public H\&E whole slides across 34 major tissue types. Evaluated on the established benchmark, GPFM achieves an impressive average rank of 1.36, with 29 tasks ranked 1st, while the the second-best model, UNI, attains an average rank of 2.96, with only 4 tasks ranked 1st. The superior generalization of GPFM demonstrates its exceptional modeling capabilities across a wide range of clinical tasks, positioning it as a new cornerstone for feature representation in CPath.
A Multimodal Knowledge-enhanced Whole-slide Pathology Foundation Model
Jul 22, 2024
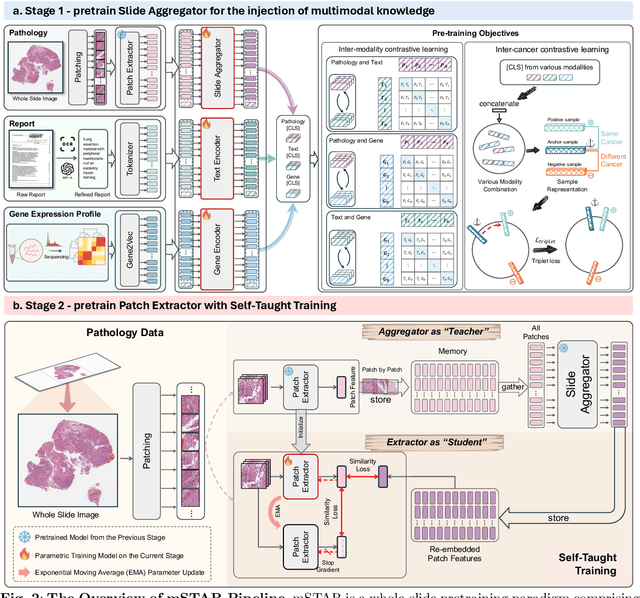
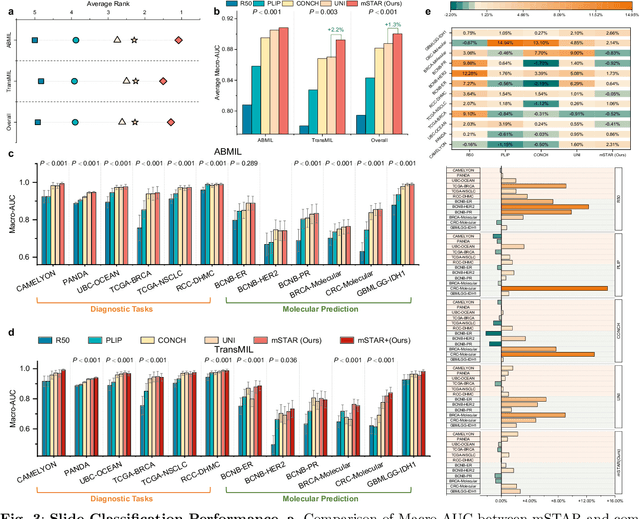

Abstract:Remarkable strides in computational pathology have been made in the task-agnostic foundation model that advances the performance of a wide array of downstream clinical tasks. Despite the promising performance, there are still several challenges. First, prior works have resorted to either vision-only or vision-captions data, disregarding invaluable pathology reports and gene expression profiles which respectively offer distinct knowledge for versatile clinical applications. Second, the current progress in pathology FMs predominantly concentrates on the patch level, where the restricted context of patch-level pretraining fails to capture whole-slide patterns. Here we curated the largest multimodal dataset consisting of H\&E diagnostic whole slide images and their associated pathology reports and RNA-Seq data, resulting in 26,169 slide-level modality pairs from 10,275 patients across 32 cancer types. To leverage these data for CPath, we propose a novel whole-slide pretraining paradigm which injects multimodal knowledge at the whole-slide context into the pathology FM, called Multimodal Self-TAught PRetraining (mSTAR). The proposed paradigm revolutionizes the workflow of pretraining for CPath, which enables the pathology FM to acquire the whole-slide context. To our knowledge, this is the first attempt to incorporate multimodal knowledge at the slide level for enhancing pathology FMs, expanding the modelling context from unimodal to multimodal knowledge and from patch-level to slide-level. To systematically evaluate the capabilities of mSTAR, extensive experiments including slide-level unimodal and multimodal applications, are conducted across 7 diverse types of tasks on 43 subtasks, resulting in the largest spectrum of downstream tasks. The average performance in various slide-level applications consistently demonstrates significant performance enhancements for mSTAR compared to SOTA FMs.
Foundation Model for Advancing Healthcare: Challenges, Opportunities, and Future Directions
Apr 04, 2024



Abstract:Foundation model, which is pre-trained on broad data and is able to adapt to a wide range of tasks, is advancing healthcare. It promotes the development of healthcare artificial intelligence (AI) models, breaking the contradiction between limited AI models and diverse healthcare practices. Much more widespread healthcare scenarios will benefit from the development of a healthcare foundation model (HFM), improving their advanced intelligent healthcare services. Despite the impending widespread deployment of HFMs, there is currently a lack of clear understanding about how they work in the healthcare field, their current challenges, and where they are headed in the future. To answer these questions, a comprehensive and deep survey of the challenges, opportunities, and future directions of HFMs is presented in this survey. It first conducted a comprehensive overview of the HFM including the methods, data, and applications for a quick grasp of the current progress. Then, it made an in-depth exploration of the challenges present in data, algorithms, and computing infrastructures for constructing and widespread application of foundation models in healthcare. This survey also identifies emerging and promising directions in this field for future development. We believe that this survey will enhance the community's comprehension of the current progress of HFM and serve as a valuable source of guidance for future development in this field. The latest HFM papers and related resources are maintained on our website: https://github.com/YutingHe-list/Awesome-Foundation-Models-for-Advancing-Healthcare.
iMD4GC: Incomplete Multimodal Data Integration to Advance Precise Treatment Response Prediction and Survival Analysis for Gastric Cancer
Apr 01, 2024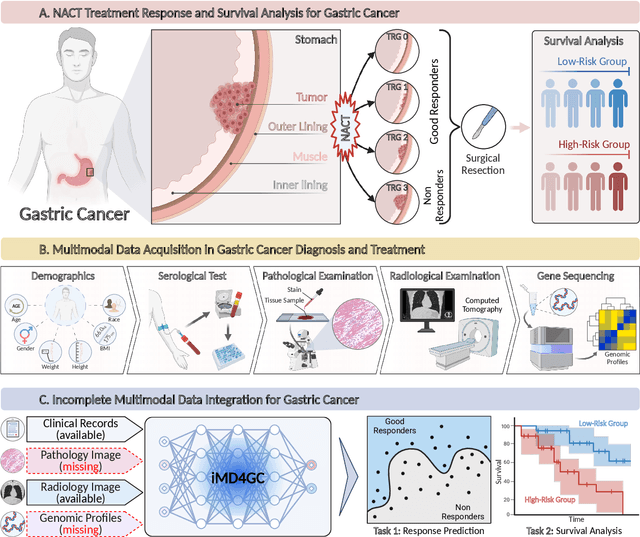
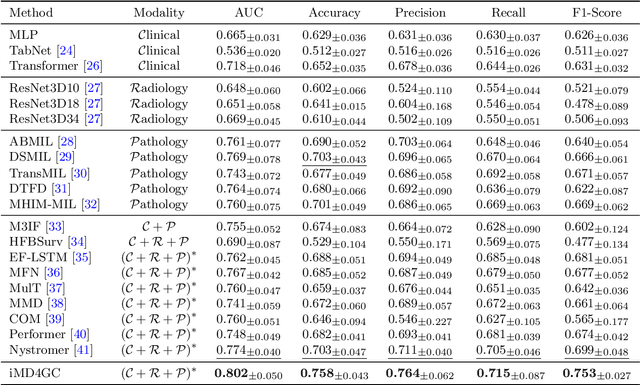
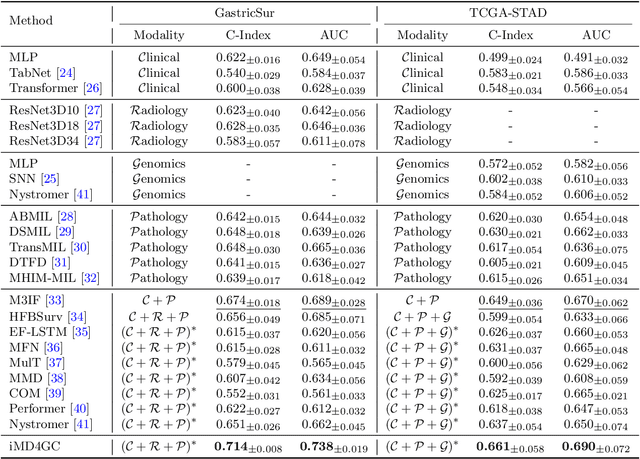
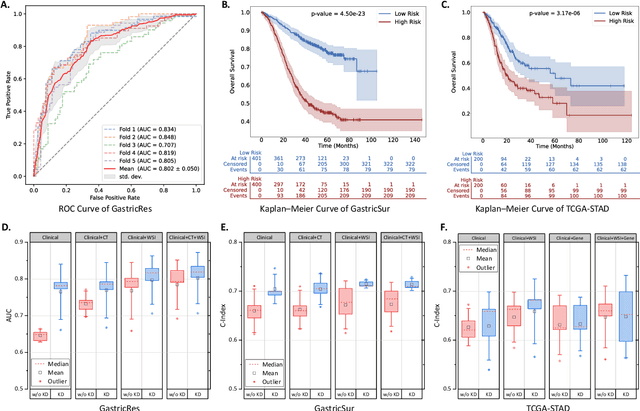
Abstract:Gastric cancer (GC) is a prevalent malignancy worldwide, ranking as the fifth most common cancer with over 1 million new cases and 700 thousand deaths in 2020. Locally advanced gastric cancer (LAGC) accounts for approximately two-thirds of GC diagnoses, and neoadjuvant chemotherapy (NACT) has emerged as the standard treatment for LAGC. However, the effectiveness of NACT varies significantly among patients, with a considerable subset displaying treatment resistance. Ineffective NACT not only leads to adverse effects but also misses the optimal therapeutic window, resulting in lower survival rate. However, existing multimodal learning methods assume the availability of all modalities for each patient, which does not align with the reality of clinical practice. The limited availability of modalities for each patient would cause information loss, adversely affecting predictive accuracy. In this study, we propose an incomplete multimodal data integration framework for GC (iMD4GC) to address the challenges posed by incomplete multimodal data, enabling precise response prediction and survival analysis. Specifically, iMD4GC incorporates unimodal attention layers for each modality to capture intra-modal information. Subsequently, the cross-modal interaction layers explore potential inter-modal interactions and capture complementary information across modalities, thereby enabling information compensation for missing modalities. To evaluate iMD4GC, we collected three multimodal datasets for GC study: GastricRes (698 cases) for response prediction, GastricSur (801 cases) for survival analysis, and TCGA-STAD (400 cases) for survival analysis. The scale of our datasets is significantly larger than previous studies. The iMD4GC achieved impressive performance with an 80.2% AUC on GastricRes, 71.4% C-index on GastricSur, and 66.1% C-index on TCGA-STAD, significantly surpassing other compared methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge