Anjia Han
Cross-Modal Prototype Allocation: Unsupervised Slide Representation Learning via Patch-Text Contrast in Computational Pathology
Mar 26, 2025Abstract:With the rapid advancement of pathology foundation models (FMs), the representation learning of whole slide images (WSIs) attracts increasing attention. Existing studies develop high-quality patch feature extractors and employ carefully designed aggregation schemes to derive slide-level representations. However, mainstream weakly supervised slide representation learning methods, primarily based on multiple instance learning (MIL), are tailored to specific downstream tasks, which limits their generalizability. To address this issue, some studies explore unsupervised slide representation learning. However, these approaches focus solely on the visual modality of patches, neglecting the rich semantic information embedded in textual data. In this work, we propose ProAlign, a cross-modal unsupervised slide representation learning framework. Specifically, we leverage a large language model (LLM) to generate descriptive text for the prototype types present in a WSI, introducing patch-text contrast to construct initial prototype embeddings. Furthermore, we propose a parameter-free attention aggregation strategy that utilizes the similarity between patches and these prototypes to form unsupervised slide embeddings applicable to a wide range of downstream tasks. Extensive experiments on four public datasets show that ProAlign outperforms existing unsupervised frameworks and achieves performance comparable to some weakly supervised models.
Can We Simplify Slide-level Fine-tuning of Pathology Foundation Models?
Feb 28, 2025Abstract:The emergence of foundation models in computational pathology has transformed histopathological image analysis, with whole slide imaging (WSI) diagnosis being a core application. Traditionally, weakly supervised fine-tuning via multiple instance learning (MIL) has been the primary method for adapting foundation models to WSIs. However, in this work we present a key experimental finding: a simple nonlinear mapping strategy combining mean pooling and a multilayer perceptron, called SiMLP, can effectively adapt patch-level foundation models to slide-level tasks without complex MIL-based learning. Through extensive experiments across diverse downstream tasks, we demonstrate the superior performance of SiMLP with state-of-the-art methods. For instance, on a large-scale pan-cancer classification task, SiMLP surpasses popular MIL-based methods by 3.52%. Furthermore, SiMLP shows strong learning ability in few-shot classification and remaining highly competitive with slide-level foundation models pretrained on tens of thousands of slides. Finally, SiMLP exhibits remarkable robustness and transferability in lung cancer subtyping. Overall, our findings challenge the conventional MIL-based fine-tuning paradigm, demonstrating that a task-agnostic representation strategy alone can effectively adapt foundation models to WSI analysis. These insights offer a unique and meaningful perspective for future research in digital pathology, paving the way for more efficient and broadly applicable methodologies.
Dynamic Hypergraph Representation for Bone Metastasis Cancer Analysis
Jan 28, 2025Abstract:Bone metastasis analysis is a significant challenge in pathology and plays a critical role in determining patient quality of life and treatment strategies. The microenvironment and specific tissue structures are essential for pathologists to predict the primary bone cancer origins and primary bone cancer subtyping. By digitizing bone tissue sections into whole slide images (WSIs) and leveraging deep learning to model slide embeddings, this analysis can be enhanced. However, tumor metastasis involves complex multivariate interactions with diverse bone tissue structures, which traditional WSI analysis methods such as multiple instance learning (MIL) fail to capture. Moreover, graph neural networks (GNNs), limited to modeling pairwise relationships, are hard to represent high-order biological associations. To address these challenges, we propose a dynamic hypergraph neural network (DyHG) that overcomes the edge construction limitations of traditional graph representations by connecting multiple nodes via hyperedges. A low-rank strategy is used to reduce the complexity of parameters in learning hypergraph structures, while a Gumbel-Softmax-based sampling strategy optimizes the patch distribution across hyperedges. An MIL aggregator is then used to derive a graph-level embedding for comprehensive WSI analysis. To evaluate the effectiveness of DyHG, we construct two large-scale datasets for primary bone cancer origins and subtyping classification based on real-world bone metastasis scenarios. Extensive experiments demonstrate that DyHG significantly outperforms state-of-the-art (SOTA) baselines, showcasing its ability to model complex biological interactions and improve the accuracy of bone metastasis analysis.
Diagnostic Text-guided Representation Learning in Hierarchical Classification for Pathological Whole Slide Image
Nov 16, 2024



Abstract:With the development of digital imaging in medical microscopy, artificial intelligent-based analysis of pathological whole slide images (WSIs) provides a powerful tool for cancer diagnosis. Limited by the expensive cost of pixel-level annotation, current research primarily focuses on representation learning with slide-level labels, showing success in various downstream tasks. However, given the diversity of lesion types and the complex relationships between each other, these techniques still deserve further exploration in addressing advanced pathology tasks. To this end, we introduce the concept of hierarchical pathological image classification and propose a representation learning called PathTree. PathTree considers the multi-classification of diseases as a binary tree structure. Each category is represented as a professional pathological text description, which messages information with a tree-like encoder. The interactive text features are then used to guide the aggregation of hierarchical multiple representations. PathTree uses slide-text similarity to obtain probability scores and introduces two extra tree specific losses to further constrain the association between texts and slides. Through extensive experiments on three challenging hierarchical classification datasets: in-house cryosectioned lung tissue lesion identification, public prostate cancer grade assessment, and public breast cancer subtyping, our proposed PathTree is consistently competitive compared to the state-of-the-art methods and provides a new perspective on the deep learning-assisted solution for more complex WSI classification.
Towards A Generalizable Pathology Foundation Model via Unified Knowledge Distillation
Jul 26, 2024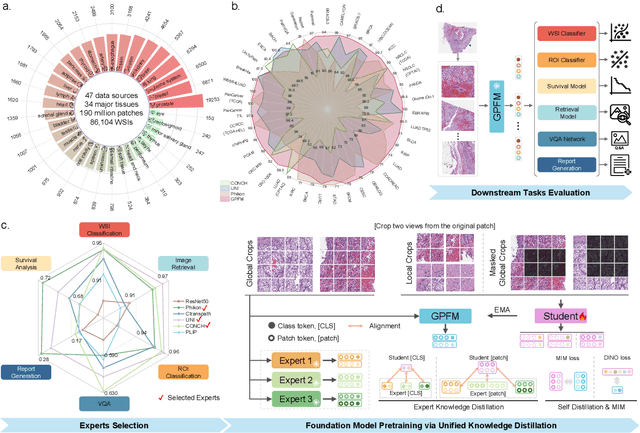

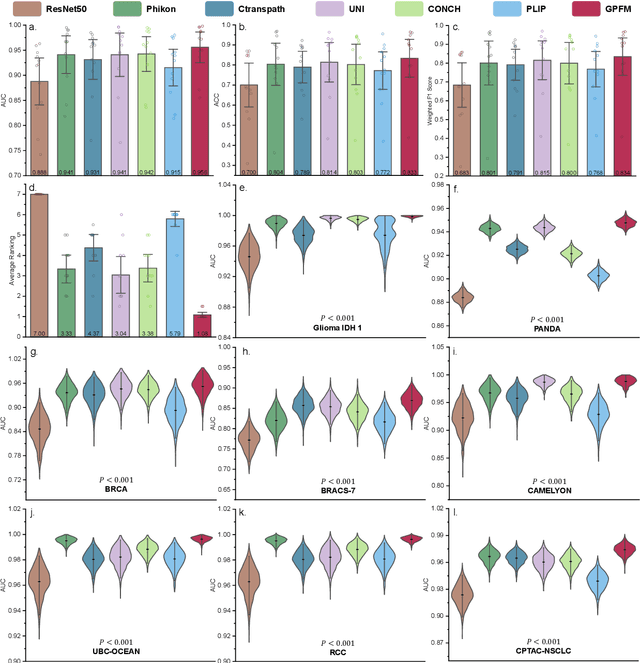
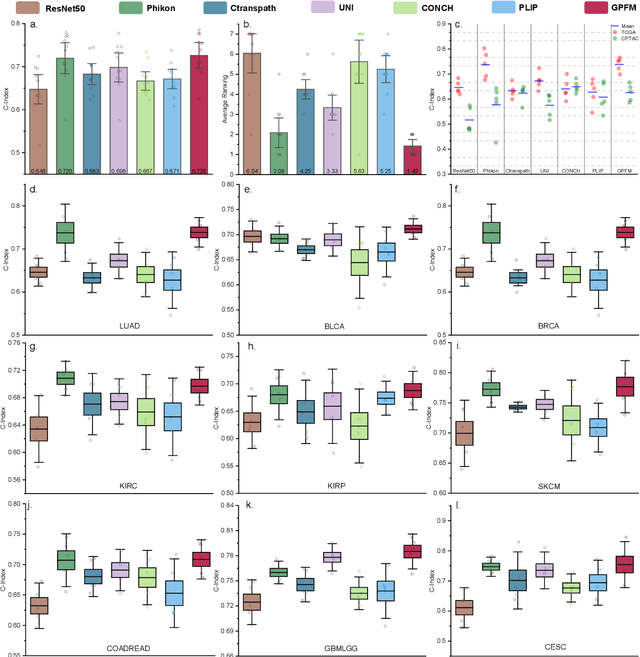
Abstract:Foundation models pretrained on large-scale datasets are revolutionizing the field of computational pathology (CPath). The generalization ability of foundation models is crucial for the success in various downstream clinical tasks. However, current foundation models have only been evaluated on a limited type and number of tasks, leaving their generalization ability and overall performance unclear. To address this gap, we established a most comprehensive benchmark to evaluate the performance of off-the-shelf foundation models across six distinct clinical task types, encompassing a total of 39 specific tasks. Our findings reveal that existing foundation models excel at certain task types but struggle to effectively handle the full breadth of clinical tasks. To improve the generalization of pathology foundation models, we propose a unified knowledge distillation framework consisting of both expert and self knowledge distillation, where the former allows the model to learn from the knowledge of multiple expert models, while the latter leverages self-distillation to enable image representation learning via local-global alignment. Based on this framework, a Generalizable Pathology Foundation Model (GPFM) is pretrained on a large-scale dataset consisting of 190 million images from around 86,000 public H\&E whole slides across 34 major tissue types. Evaluated on the established benchmark, GPFM achieves an impressive average rank of 1.36, with 29 tasks ranked 1st, while the the second-best model, UNI, attains an average rank of 2.96, with only 4 tasks ranked 1st. The superior generalization of GPFM demonstrates its exceptional modeling capabilities across a wide range of clinical tasks, positioning it as a new cornerstone for feature representation in CPath.
A Multimodal Knowledge-enhanced Whole-slide Pathology Foundation Model
Jul 22, 2024
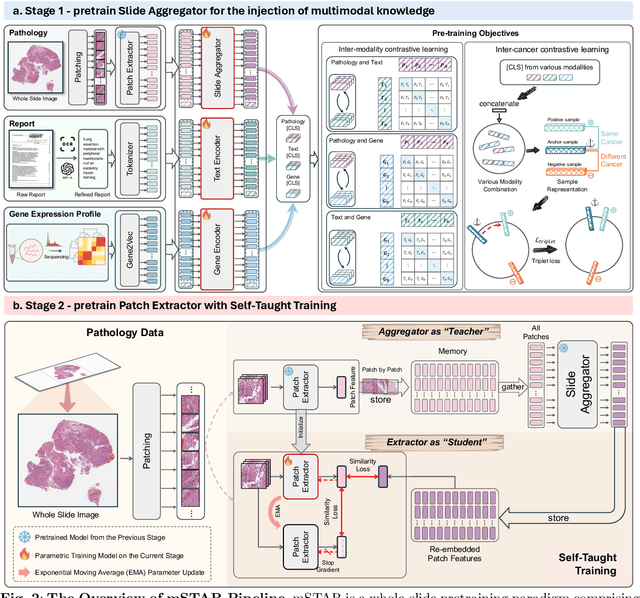
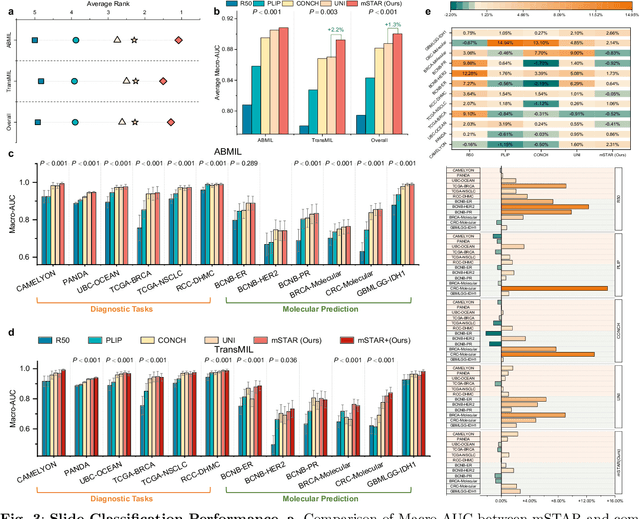

Abstract:Remarkable strides in computational pathology have been made in the task-agnostic foundation model that advances the performance of a wide array of downstream clinical tasks. Despite the promising performance, there are still several challenges. First, prior works have resorted to either vision-only or vision-captions data, disregarding invaluable pathology reports and gene expression profiles which respectively offer distinct knowledge for versatile clinical applications. Second, the current progress in pathology FMs predominantly concentrates on the patch level, where the restricted context of patch-level pretraining fails to capture whole-slide patterns. Here we curated the largest multimodal dataset consisting of H\&E diagnostic whole slide images and their associated pathology reports and RNA-Seq data, resulting in 26,169 slide-level modality pairs from 10,275 patients across 32 cancer types. To leverage these data for CPath, we propose a novel whole-slide pretraining paradigm which injects multimodal knowledge at the whole-slide context into the pathology FM, called Multimodal Self-TAught PRetraining (mSTAR). The proposed paradigm revolutionizes the workflow of pretraining for CPath, which enables the pathology FM to acquire the whole-slide context. To our knowledge, this is the first attempt to incorporate multimodal knowledge at the slide level for enhancing pathology FMs, expanding the modelling context from unimodal to multimodal knowledge and from patch-level to slide-level. To systematically evaluate the capabilities of mSTAR, extensive experiments including slide-level unimodal and multimodal applications, are conducted across 7 diverse types of tasks on 43 subtasks, resulting in the largest spectrum of downstream tasks. The average performance in various slide-level applications consistently demonstrates significant performance enhancements for mSTAR compared to SOTA FMs.
Leveraging Pre-trained Models for FF-to-FFPE Histopathological Image Translation
Jun 26, 2024



Abstract:The two primary types of Hematoxylin and Eosin (H&E) slides in histopathology are Formalin-Fixed Paraffin-Embedded (FFPE) and Fresh Frozen (FF). FFPE slides offer high quality histopathological images but require a labor-intensive acquisition process. In contrast, FF slides can be prepared quickly, but the image quality is relatively poor. Our task is to translate FF images into FFPE style, thereby improving the image quality for diagnostic purposes. In this paper, we propose Diffusion-FFPE, a method for FF-to-FFPE histopathological image translation using a pre-trained diffusion model. Specifically, we employ a one-step diffusion model as the generator and fine-tune it with LoRA adapters using adversarial learning objectives. To ensure that the model effectively captures both global structural information and local details, we propose a multi-scale feature fusion (MFF) module. This module utilizes two VAE encoders to extract features of varying image sizes and performs feature fusion before feeding them into the UNet. Furthermore, we utilize a pre-trained vision-language model for histopathology as the backbone for the discriminator to further improve performance We conducted FF-to-FFPE translation experiments on the TCGA-NSCLC datasets, and our method achieved better performance compared to other methods. The code and models are released at https://github.com/QilaiZhang/Diffusion-FFPE.
RetMIL: Retentive Multiple Instance Learning for Histopathological Whole Slide Image Classification
Mar 16, 2024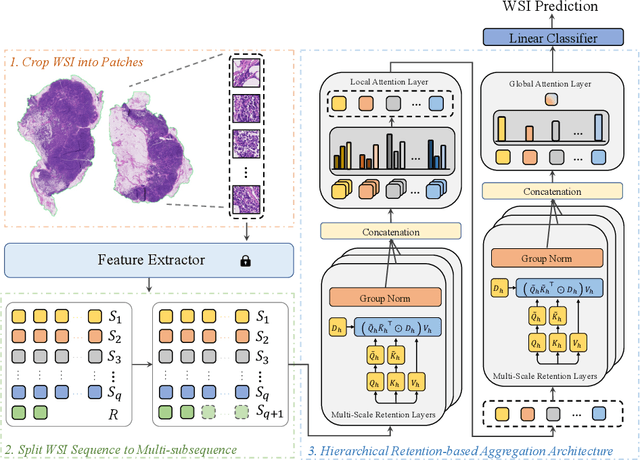
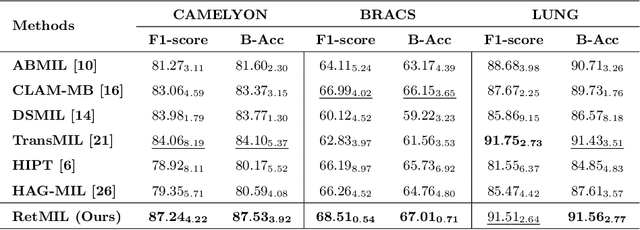
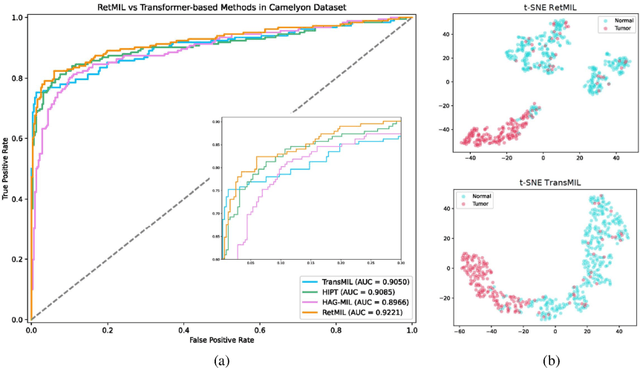
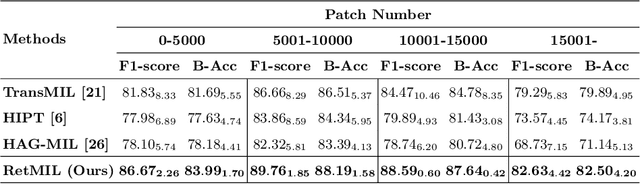
Abstract:Histopathological whole slide image (WSI) analysis with deep learning has become a research focus in computational pathology. The current paradigm is mainly based on multiple instance learning (MIL), in which approaches with Transformer as the backbone are well discussed. These methods convert WSI tasks into sequence tasks by representing patches as tokens in the WSI sequence. However, the feature complexity brought by high heterogeneity and the ultra-long sequences brought by gigapixel size makes Transformer-based MIL suffer from the challenges of high memory consumption, slow inference speed, and lack of performance. To this end, we propose a retentive MIL method called RetMIL, which processes WSI sequences through hierarchical feature propagation structure. At the local level, the WSI sequence is divided into multiple subsequences. Tokens of each subsequence are updated through a parallel linear retention mechanism and aggregated utilizing an attention layer. At the global level, subsequences are fused into a global sequence, then updated through a serial retention mechanism, and finally the slide-level representation is obtained through a global attention pooling. We conduct experiments on two public CAMELYON and BRACS datasets and an public-internal LUNG dataset, confirming that RetMIL not only achieves state-of-the-art performance but also significantly reduces computational overhead. Our code will be accessed shortly.
Dynamic Graph Representation with Knowledge-aware Attention for Histopathology Whole Slide Image Analysis
Mar 12, 2024



Abstract:Histopathological whole slide images (WSIs) classification has become a foundation task in medical microscopic imaging processing. Prevailing approaches involve learning WSIs as instance-bag representations, emphasizing significant instances but struggling to capture the interactions between instances. Additionally, conventional graph representation methods utilize explicit spatial positions to construct topological structures but restrict the flexible interaction capabilities between instances at arbitrary locations, particularly when spatially distant. In response, we propose a novel dynamic graph representation algorithm that conceptualizes WSIs as a form of the knowledge graph structure. Specifically, we dynamically construct neighbors and directed edge embeddings based on the head and tail relationships between instances. Then, we devise a knowledge-aware attention mechanism that can update the head node features by learning the joint attention score of each neighbor and edge. Finally, we obtain a graph-level embedding through the global pooling process of the updated head, serving as an implicit representation for the WSI classification. Our end-to-end graph representation learning approach has outperformed the state-of-the-art WSI analysis methods on three TCGA benchmark datasets and in-house test sets. Our code is available at https://github.com/WonderLandxD/WiKG.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge