Tian Guan
To What Extent Do Token-Level Representations from Pathology Foundation Models Improve Dense Prediction?
Feb 03, 2026Abstract:Pathology foundation models (PFMs) have rapidly advanced and are becoming a common backbone for downstream clinical tasks, offering strong transferability across tissues and institutions. However, for dense prediction (e.g., segmentation), practical deployment still lacks a clear, reproducible understanding of how different PFMs behave across datasets and how adaptation choices affect performance and stability. We present PFM-DenseBench, a large-scale benchmark for dense pathology prediction, evaluating 17 PFMs across 18 public segmentation datasets. Under a unified protocol, we systematically assess PFMs with multiple adaptation and fine-tuning strategies, and derive insightful, practice-oriented findings on when and why different PFMs and tuning choices succeed or fail across heterogeneous datasets. We release containers, configs, and dataset cards to enable reproducible evaluation and informed PFM selection for real-world dense pathology tasks. Project Website: https://m4a1tastegood.github.io/PFM-DenseBench
HookMIL: Revisiting Context Modeling in Multiple Instance Learning for Computational Pathology
Dec 20, 2025Abstract:Multiple Instance Learning (MIL) has enabled weakly supervised analysis of whole-slide images (WSIs) in computational pathology. However, traditional MIL approaches often lose crucial contextual information, while transformer-based variants, though more expressive, suffer from quadratic complexity and redundant computations. To address these limitations, we propose HookMIL, a context-aware and computationally efficient MIL framework that leverages compact, learnable hook tokens for structured contextual aggregation. These tokens can be initialized from (i) key-patch visual features, (ii) text embeddings from vision-language pathology models, and (iii) spatially grounded features from spatial transcriptomics-vision models. This multimodal initialization enables Hook Tokens to incorporate rich textual and spatial priors, accelerating convergence and enhancing representation quality. During training, Hook tokens interact with instances through bidirectional attention with linear complexity. To further promote specialization, we introduce a Hook Diversity Loss that encourages each token to focus on distinct histopathological patterns. Additionally, a hook-to-hook communication mechanism refines contextual interactions while minimizing redundancy. Extensive experiments on four public pathology datasets demonstrate that HookMIL achieves state-of-the-art performance, with improved computational efficiency and interpretability. Codes are available at https://github.com/lingxitong/HookMIL.
StainNet: A Special Staining Self-Supervised Vision Transformer for Computational Pathology
Dec 11, 2025Abstract:Foundation models trained with self-supervised learning (SSL) on large-scale histological images have significantly accelerated the development of computational pathology. These models can serve as backbones for region-of-interest (ROI) image analysis or patch-level feature extractors in whole-slide images (WSIs) based on multiple instance learning (MIL). Existing pathology foundation models (PFMs) are typically pre-trained on Hematoxylin-Eosin (H&E) stained pathology images. However, images with special stains, such as immunohistochemistry, are also frequently used in clinical practice. PFMs pre-trained mainly on H\&E-stained images may be limited in clinical applications involving special stains. To address this issue, we propose StainNet, a specialized foundation model for special stains based on the vision transformer (ViT) architecture. StainNet adopts a self-distillation SSL approach and is trained on over 1.4 million patch images cropping from 20,231 publicly available special staining WSIs in the HISTAI database. To evaluate StainNet, we conduct experiments on an in-house slide-level liver malignancy classification task and two public ROI-level datasets to demonstrate its strong ability. We also perform few-ratio learning and retrieval evaluations, and compare StainNet with recently larger PFMs to further highlight its strengths. We have released the StainNet model weights at: https://huggingface.co/JWonderLand/StainNet.
Multimodal Prototype Alignment for Semi-supervised Pathology Image Segmentation
Aug 27, 2025Abstract:Pathological image segmentation faces numerous challenges, particularly due to ambiguous semantic boundaries and the high cost of pixel-level annotations. Although recent semi-supervised methods based on consistency regularization (e.g., UniMatch) have made notable progress, they mainly rely on perturbation-based consistency within the image modality, making it difficult to capture high-level semantic priors, especially in structurally complex pathology images. To address these limitations, we propose MPAMatch - a novel segmentation framework that performs pixel-level contrastive learning under a multimodal prototype-guided supervision paradigm. The core innovation of MPAMatch lies in the dual contrastive learning scheme between image prototypes and pixel labels, and between text prototypes and pixel labels, providing supervision at both structural and semantic levels. This coarse-to-fine supervisory strategy not only enhances the discriminative capability on unlabeled samples but also introduces the text prototype supervision into segmentation for the first time, significantly improving semantic boundary modeling. In addition, we reconstruct the classic segmentation architecture (TransUNet) by replacing its ViT backbone with a pathology-pretrained foundation model (Uni), enabling more effective extraction of pathology-relevant features. Extensive experiments on GLAS, EBHI-SEG-GLAND, EBHI-SEG-CANCER, and KPI show MPAMatch's superiority over state-of-the-art methods, validating its dual advantages in structural and semantic modeling.
Deformable Attention Graph Representation Learning for Histopathology Whole Slide Image Analysis
Aug 07, 2025Abstract:Accurate classification of Whole Slide Images (WSIs) and Regions of Interest (ROIs) is a fundamental challenge in computational pathology. While mainstream approaches often adopt Multiple Instance Learning (MIL), they struggle to capture the spatial dependencies among tissue structures. Graph Neural Networks (GNNs) have emerged as a solution to model inter-instance relationships, yet most rely on static graph topologies and overlook the physical spatial positions of tissue patches. Moreover, conventional attention mechanisms lack specificity, limiting their ability to focus on structurally relevant regions. In this work, we propose a novel GNN framework with deformable attention for pathology image analysis. We construct a dynamic weighted directed graph based on patch features, where each node aggregates contextual information from its neighbors via attention-weighted edges. Specifically, we incorporate learnable spatial offsets informed by the real coordinates of each patch, enabling the model to adaptively attend to morphologically relevant regions across the slide. This design significantly enhances the contextual field while preserving spatial specificity. Our framework achieves state-of-the-art performance on four benchmark datasets (TCGA-COAD, BRACS, gastric intestinal metaplasia grading, and intestinal ROI classification), demonstrating the power of deformable attention in capturing complex spatial structures in WSIs and ROIs.
A Simple Linear Patch Revives Layer-Pruned Large Language Models
May 30, 2025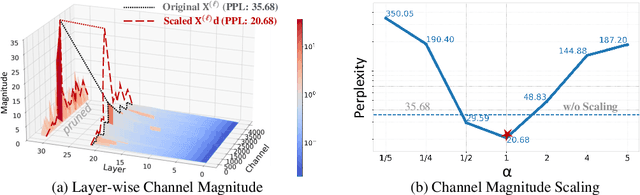
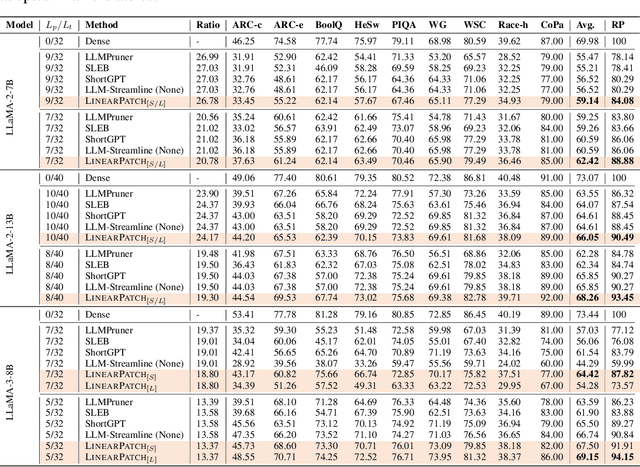
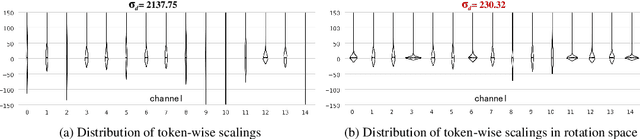
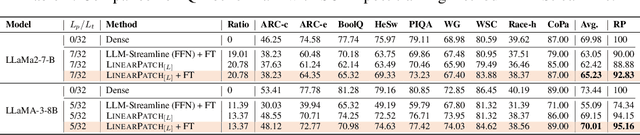
Abstract:Layer pruning has become a popular technique for compressing large language models (LLMs) due to its simplicity. However, existing layer pruning methods often suffer from significant performance drops. We identify that this degradation stems from the mismatch of activation magnitudes across layers and tokens at the pruning interface. To address this, we propose LinearPatch, a simple plug-and-play technique to revive the layer-pruned LLMs. The proposed method adopts Hadamard transformation to suppress massive outliers in particular tokens, and channel-wise scaling to align the activation magnitudes. These operations can be fused into a single matrix, which functions as a patch to bridge the pruning interface with negligible inference overhead. LinearPatch retains up to 94.15% performance of the original model when pruning 5 layers of LLaMA-3-8B on the question answering benchmark, surpassing existing state-of-the-art methods by 4%. In addition, the patch matrix can be further optimized with memory efficient offline knowledge distillation. With only 5K samples, the retained performance of LinearPatch can be further boosted to 95.16% within 30 minutes on a single computing card.
Subspecialty-Specific Foundation Model for Intelligent Gastrointestinal Pathology
May 28, 2025Abstract:Gastrointestinal (GI) diseases represent a clinically significant burden, necessitating precise diagnostic approaches to optimize patient outcomes. Conventional histopathological diagnosis, heavily reliant on the subjective interpretation of pathologists, suffers from limited reproducibility and diagnostic variability. To overcome these limitations and address the lack of pathology-specific foundation models for GI diseases, we develop Digepath, a specialized foundation model for GI pathology. Our framework introduces a dual-phase iterative optimization strategy combining pretraining with fine-screening, specifically designed to address the detection of sparsely distributed lesion areas in whole-slide images. Digepath is pretrained on more than 353 million image patches from over 200,000 hematoxylin and eosin-stained slides of GI diseases. It attains state-of-the-art performance on 33 out of 34 tasks related to GI pathology, including pathological diagnosis, molecular prediction, gene mutation prediction, and prognosis evaluation, particularly in diagnostically ambiguous cases and resolution-agnostic tissue classification.We further translate the intelligent screening module for early GI cancer and achieve near-perfect 99.6% sensitivity across 9 independent medical institutions nationwide. The outstanding performance of Digepath highlights its potential to bridge critical gaps in histopathological practice. This work not only advances AI-driven precision pathology for GI diseases but also establishes a transferable paradigm for other pathology subspecialties.
Cross-Modal Prototype Allocation: Unsupervised Slide Representation Learning via Patch-Text Contrast in Computational Pathology
Mar 26, 2025Abstract:With the rapid advancement of pathology foundation models (FMs), the representation learning of whole slide images (WSIs) attracts increasing attention. Existing studies develop high-quality patch feature extractors and employ carefully designed aggregation schemes to derive slide-level representations. However, mainstream weakly supervised slide representation learning methods, primarily based on multiple instance learning (MIL), are tailored to specific downstream tasks, which limits their generalizability. To address this issue, some studies explore unsupervised slide representation learning. However, these approaches focus solely on the visual modality of patches, neglecting the rich semantic information embedded in textual data. In this work, we propose ProAlign, a cross-modal unsupervised slide representation learning framework. Specifically, we leverage a large language model (LLM) to generate descriptive text for the prototype types present in a WSI, introducing patch-text contrast to construct initial prototype embeddings. Furthermore, we propose a parameter-free attention aggregation strategy that utilizes the similarity between patches and these prototypes to form unsupervised slide embeddings applicable to a wide range of downstream tasks. Extensive experiments on four public datasets show that ProAlign outperforms existing unsupervised frameworks and achieves performance comparable to some weakly supervised models.
Multimodal Distillation-Driven Ensemble Learning for Long-Tailed Histopathology Whole Slide Images Analysis
Mar 02, 2025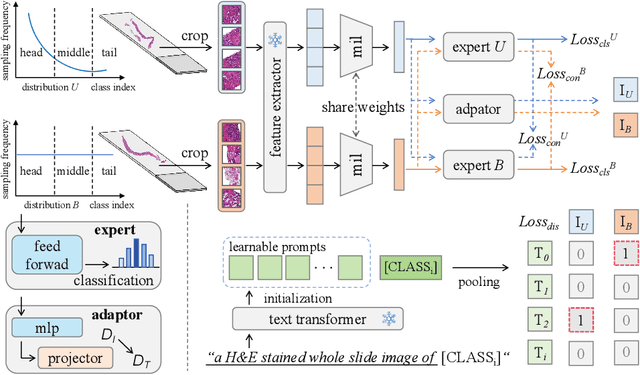


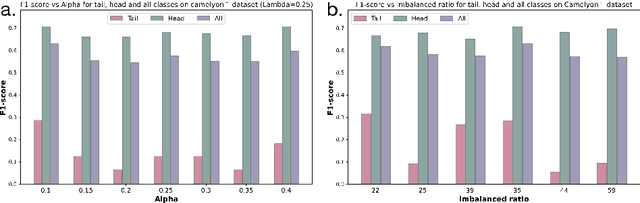
Abstract:Multiple Instance Learning (MIL) plays a significant role in computational pathology, enabling weakly supervised analysis of Whole Slide Image (WSI) datasets. The field of WSI analysis is confronted with a severe long-tailed distribution problem, which significantly impacts the performance of classifiers. Long-tailed distributions lead to class imbalance, where some classes have sparse samples while others are abundant, making it difficult for classifiers to accurately identify minority class samples. To address this issue, we propose an ensemble learning method based on MIL, which employs expert decoders with shared aggregators and consistency constraints to learn diverse distributions and reduce the impact of class imbalance on classifier performance. Moreover, we introduce a multimodal distillation framework that leverages text encoders pre-trained on pathology-text pairs to distill knowledge and guide the MIL aggregator in capturing stronger semantic features relevant to class information. To ensure flexibility, we use learnable prompts to guide the distillation process of the pre-trained text encoder, avoiding limitations imposed by specific prompts. Our method, MDE-MIL, integrates multiple expert branches focusing on specific data distributions to address long-tailed issues. Consistency control ensures generalization across classes. Multimodal distillation enhances feature extraction. Experiments on Camelyon+-LT and PANDA-LT datasets show it outperforms state-of-the-art methods.
Can We Simplify Slide-level Fine-tuning of Pathology Foundation Models?
Feb 28, 2025Abstract:The emergence of foundation models in computational pathology has transformed histopathological image analysis, with whole slide imaging (WSI) diagnosis being a core application. Traditionally, weakly supervised fine-tuning via multiple instance learning (MIL) has been the primary method for adapting foundation models to WSIs. However, in this work we present a key experimental finding: a simple nonlinear mapping strategy combining mean pooling and a multilayer perceptron, called SiMLP, can effectively adapt patch-level foundation models to slide-level tasks without complex MIL-based learning. Through extensive experiments across diverse downstream tasks, we demonstrate the superior performance of SiMLP with state-of-the-art methods. For instance, on a large-scale pan-cancer classification task, SiMLP surpasses popular MIL-based methods by 3.52%. Furthermore, SiMLP shows strong learning ability in few-shot classification and remaining highly competitive with slide-level foundation models pretrained on tens of thousands of slides. Finally, SiMLP exhibits remarkable robustness and transferability in lung cancer subtyping. Overall, our findings challenge the conventional MIL-based fine-tuning paradigm, demonstrating that a task-agnostic representation strategy alone can effectively adapt foundation models to WSI analysis. These insights offer a unique and meaningful perspective for future research in digital pathology, paving the way for more efficient and broadly applicable methodologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge