Xitong Ling
To What Extent Do Token-Level Representations from Pathology Foundation Models Improve Dense Prediction?
Feb 03, 2026Abstract:Pathology foundation models (PFMs) have rapidly advanced and are becoming a common backbone for downstream clinical tasks, offering strong transferability across tissues and institutions. However, for dense prediction (e.g., segmentation), practical deployment still lacks a clear, reproducible understanding of how different PFMs behave across datasets and how adaptation choices affect performance and stability. We present PFM-DenseBench, a large-scale benchmark for dense pathology prediction, evaluating 17 PFMs across 18 public segmentation datasets. Under a unified protocol, we systematically assess PFMs with multiple adaptation and fine-tuning strategies, and derive insightful, practice-oriented findings on when and why different PFMs and tuning choices succeed or fail across heterogeneous datasets. We release containers, configs, and dataset cards to enable reproducible evaluation and informed PFM selection for real-world dense pathology tasks. Project Website: https://m4a1tastegood.github.io/PFM-DenseBench
HookMIL: Revisiting Context Modeling in Multiple Instance Learning for Computational Pathology
Dec 20, 2025Abstract:Multiple Instance Learning (MIL) has enabled weakly supervised analysis of whole-slide images (WSIs) in computational pathology. However, traditional MIL approaches often lose crucial contextual information, while transformer-based variants, though more expressive, suffer from quadratic complexity and redundant computations. To address these limitations, we propose HookMIL, a context-aware and computationally efficient MIL framework that leverages compact, learnable hook tokens for structured contextual aggregation. These tokens can be initialized from (i) key-patch visual features, (ii) text embeddings from vision-language pathology models, and (iii) spatially grounded features from spatial transcriptomics-vision models. This multimodal initialization enables Hook Tokens to incorporate rich textual and spatial priors, accelerating convergence and enhancing representation quality. During training, Hook tokens interact with instances through bidirectional attention with linear complexity. To further promote specialization, we introduce a Hook Diversity Loss that encourages each token to focus on distinct histopathological patterns. Additionally, a hook-to-hook communication mechanism refines contextual interactions while minimizing redundancy. Extensive experiments on four public pathology datasets demonstrate that HookMIL achieves state-of-the-art performance, with improved computational efficiency and interpretability. Codes are available at https://github.com/lingxitong/HookMIL.
StainNet: A Special Staining Self-Supervised Vision Transformer for Computational Pathology
Dec 11, 2025Abstract:Foundation models trained with self-supervised learning (SSL) on large-scale histological images have significantly accelerated the development of computational pathology. These models can serve as backbones for region-of-interest (ROI) image analysis or patch-level feature extractors in whole-slide images (WSIs) based on multiple instance learning (MIL). Existing pathology foundation models (PFMs) are typically pre-trained on Hematoxylin-Eosin (H&E) stained pathology images. However, images with special stains, such as immunohistochemistry, are also frequently used in clinical practice. PFMs pre-trained mainly on H\&E-stained images may be limited in clinical applications involving special stains. To address this issue, we propose StainNet, a specialized foundation model for special stains based on the vision transformer (ViT) architecture. StainNet adopts a self-distillation SSL approach and is trained on over 1.4 million patch images cropping from 20,231 publicly available special staining WSIs in the HISTAI database. To evaluate StainNet, we conduct experiments on an in-house slide-level liver malignancy classification task and two public ROI-level datasets to demonstrate its strong ability. We also perform few-ratio learning and retrieval evaluations, and compare StainNet with recently larger PFMs to further highlight its strengths. We have released the StainNet model weights at: https://huggingface.co/JWonderLand/StainNet.
Deep Pathomic Learning Defines Prognostic Subtypes and Molecular Drivers in Colorectal Cancer
Nov 19, 2025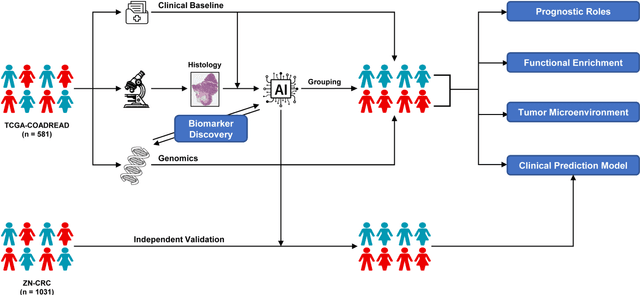
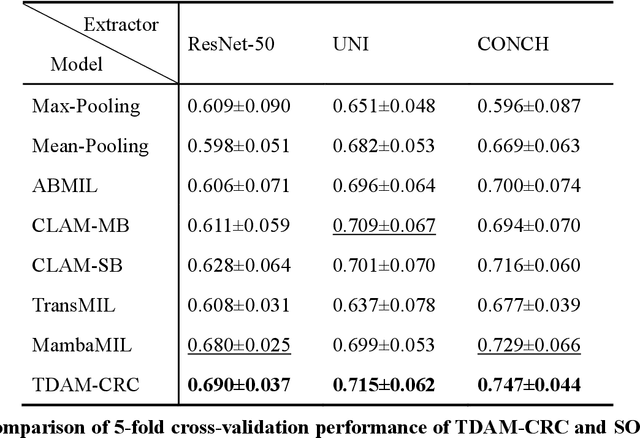
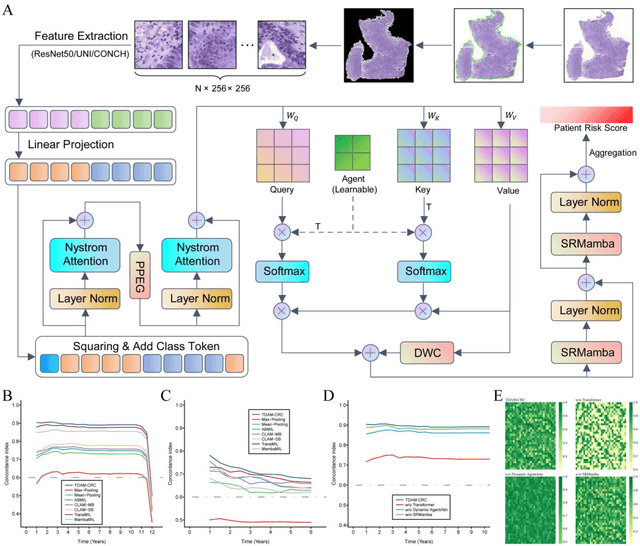
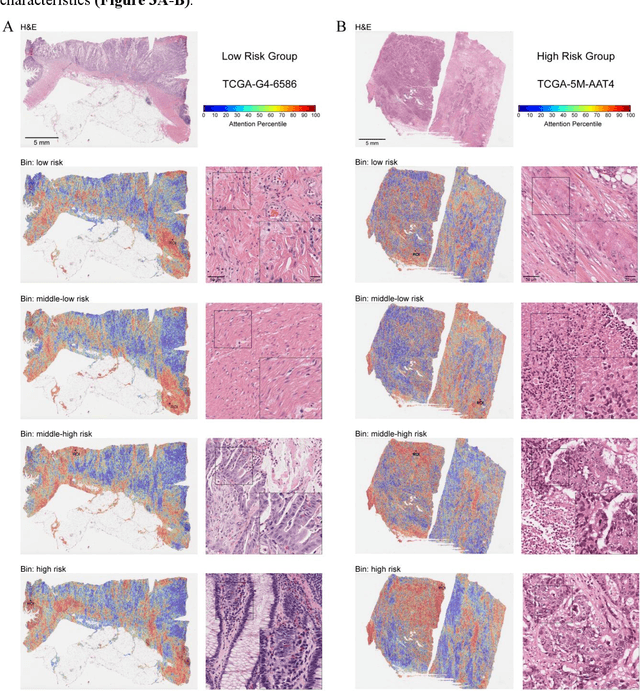
Abstract:Precise prognostic stratification of colorectal cancer (CRC) remains a major clinical challenge due to its high heterogeneity. The conventional TNM staging system is inadequate for personalized medicine. We aimed to develop and validate a novel multiple instance learning model TDAM-CRC using histopathological whole-slide images for accurate prognostic prediction and to uncover its underlying molecular mechanisms. We trained the model on the TCGA discovery cohort (n=581), validated it in an independent external cohort (n=1031), and further we integrated multi-omics data to improve model interpretability and identify novel prognostic biomarkers. The results demonstrated that the TDAM-CRC achieved robust risk stratification in both cohorts. Its predictive performance significantly outperformed the conventional clinical staging system and multiple state-of-the-art models. The TDAM-CRC risk score was confirmed as an independent prognostic factor in multivariable analysis. Multi-omics analysis revealed that the high-risk subtype is closely associated with metabolic reprogramming and an immunosuppressive tumor microenvironment. Through interaction network analysis, we identified and validated Mitochondrial Ribosomal Protein L37 (MRPL37) as a key hub gene linking deep pathomic features to clinical prognosis. We found that high expression of MRPL37, driven by promoter hypomethylation, serves as an independent biomarker of favorable prognosis. Finally, we constructed a nomogram incorporating the TDAM-CRC risk score and clinical factors to provide a precise and interpretable clinical decision-making tool for CRC patients. Our AI-driven pathological model TDAM-CRC provides a robust tool for improved CRC risk stratification, reveals new molecular targets, and facilitates personalized clinical decision-making.
SAC-MIL: Spatial-Aware Correlated Multiple Instance Learning for Histopathology Whole Slide Image Classification
Sep 04, 2025



Abstract:We propose Spatial-Aware Correlated Multiple Instance Learning (SAC-MIL) for performing WSI classification. SAC-MIL consists of a positional encoding module to encode position information and a SAC block to perform full instance correlations. The positional encoding module utilizes the instance coordinates within the slide to encode the spatial relationships instead of the instance index in the input WSI sequence. The positional encoding module can also handle the length extrapolation issue where the training and testing sequences have different lengths. The SAC block is an MLP-based method that performs full instance correlation in linear time complexity with respect to the sequence length. Due to the simple structure of MLP, it is easy to deploy since it does not require custom CUDA kernels, compared to Transformer-based methods for WSI classification. SAC-MIL has achieved state-of-the-art performance on the CAMELYON-16, TCGA-LUNG, and TCGA-BRAC datasets. The code will be released upon acceptance.
Multimodal Prototype Alignment for Semi-supervised Pathology Image Segmentation
Aug 27, 2025Abstract:Pathological image segmentation faces numerous challenges, particularly due to ambiguous semantic boundaries and the high cost of pixel-level annotations. Although recent semi-supervised methods based on consistency regularization (e.g., UniMatch) have made notable progress, they mainly rely on perturbation-based consistency within the image modality, making it difficult to capture high-level semantic priors, especially in structurally complex pathology images. To address these limitations, we propose MPAMatch - a novel segmentation framework that performs pixel-level contrastive learning under a multimodal prototype-guided supervision paradigm. The core innovation of MPAMatch lies in the dual contrastive learning scheme between image prototypes and pixel labels, and between text prototypes and pixel labels, providing supervision at both structural and semantic levels. This coarse-to-fine supervisory strategy not only enhances the discriminative capability on unlabeled samples but also introduces the text prototype supervision into segmentation for the first time, significantly improving semantic boundary modeling. In addition, we reconstruct the classic segmentation architecture (TransUNet) by replacing its ViT backbone with a pathology-pretrained foundation model (Uni), enabling more effective extraction of pathology-relevant features. Extensive experiments on GLAS, EBHI-SEG-GLAND, EBHI-SEG-CANCER, and KPI show MPAMatch's superiority over state-of-the-art methods, validating its dual advantages in structural and semantic modeling.
Deformable Attention Graph Representation Learning for Histopathology Whole Slide Image Analysis
Aug 07, 2025Abstract:Accurate classification of Whole Slide Images (WSIs) and Regions of Interest (ROIs) is a fundamental challenge in computational pathology. While mainstream approaches often adopt Multiple Instance Learning (MIL), they struggle to capture the spatial dependencies among tissue structures. Graph Neural Networks (GNNs) have emerged as a solution to model inter-instance relationships, yet most rely on static graph topologies and overlook the physical spatial positions of tissue patches. Moreover, conventional attention mechanisms lack specificity, limiting their ability to focus on structurally relevant regions. In this work, we propose a novel GNN framework with deformable attention for pathology image analysis. We construct a dynamic weighted directed graph based on patch features, where each node aggregates contextual information from its neighbors via attention-weighted edges. Specifically, we incorporate learnable spatial offsets informed by the real coordinates of each patch, enabling the model to adaptively attend to morphologically relevant regions across the slide. This design significantly enhances the contextual field while preserving spatial specificity. Our framework achieves state-of-the-art performance on four benchmark datasets (TCGA-COAD, BRACS, gastric intestinal metaplasia grading, and intestinal ROI classification), demonstrating the power of deformable attention in capturing complex spatial structures in WSIs and ROIs.
Subspecialty-Specific Foundation Model for Intelligent Gastrointestinal Pathology
May 28, 2025Abstract:Gastrointestinal (GI) diseases represent a clinically significant burden, necessitating precise diagnostic approaches to optimize patient outcomes. Conventional histopathological diagnosis, heavily reliant on the subjective interpretation of pathologists, suffers from limited reproducibility and diagnostic variability. To overcome these limitations and address the lack of pathology-specific foundation models for GI diseases, we develop Digepath, a specialized foundation model for GI pathology. Our framework introduces a dual-phase iterative optimization strategy combining pretraining with fine-screening, specifically designed to address the detection of sparsely distributed lesion areas in whole-slide images. Digepath is pretrained on more than 353 million image patches from over 200,000 hematoxylin and eosin-stained slides of GI diseases. It attains state-of-the-art performance on 33 out of 34 tasks related to GI pathology, including pathological diagnosis, molecular prediction, gene mutation prediction, and prognosis evaluation, particularly in diagnostically ambiguous cases and resolution-agnostic tissue classification.We further translate the intelligent screening module for early GI cancer and achieve near-perfect 99.6% sensitivity across 9 independent medical institutions nationwide. The outstanding performance of Digepath highlights its potential to bridge critical gaps in histopathological practice. This work not only advances AI-driven precision pathology for GI diseases but also establishes a transferable paradigm for other pathology subspecialties.
PathOrchestra: A Comprehensive Foundation Model for Computational Pathology with Over 100 Diverse Clinical-Grade Tasks
Mar 31, 2025Abstract:The complexity and variability inherent in high-resolution pathological images present significant challenges in computational pathology. While pathology foundation models leveraging AI have catalyzed transformative advancements, their development demands large-scale datasets, considerable storage capacity, and substantial computational resources. Furthermore, ensuring their clinical applicability and generalizability requires rigorous validation across a broad spectrum of clinical tasks. Here, we present PathOrchestra, a versatile pathology foundation model trained via self-supervised learning on a dataset comprising 300K pathological slides from 20 tissue and organ types across multiple centers. The model was rigorously evaluated on 112 clinical tasks using a combination of 61 private and 51 public datasets. These tasks encompass digital slide preprocessing, pan-cancer classification, lesion identification, multi-cancer subtype classification, biomarker assessment, gene expression prediction, and the generation of structured reports. PathOrchestra demonstrated exceptional performance across 27,755 WSIs and 9,415,729 ROIs, achieving over 0.950 accuracy in 47 tasks, including pan-cancer classification across various organs, lymphoma subtype diagnosis, and bladder cancer screening. Notably, it is the first model to generate structured reports for high-incidence colorectal cancer and diagnostically complex lymphoma-areas that are infrequently addressed by foundational models but hold immense clinical potential. Overall, PathOrchestra exemplifies the feasibility and efficacy of a large-scale, self-supervised pathology foundation model, validated across a broad range of clinical-grade tasks. Its high accuracy and reduced reliance on extensive data annotation underline its potential for clinical integration, offering a pathway toward more efficient and high-quality medical services.
Cross-Modal Prototype Allocation: Unsupervised Slide Representation Learning via Patch-Text Contrast in Computational Pathology
Mar 26, 2025Abstract:With the rapid advancement of pathology foundation models (FMs), the representation learning of whole slide images (WSIs) attracts increasing attention. Existing studies develop high-quality patch feature extractors and employ carefully designed aggregation schemes to derive slide-level representations. However, mainstream weakly supervised slide representation learning methods, primarily based on multiple instance learning (MIL), are tailored to specific downstream tasks, which limits their generalizability. To address this issue, some studies explore unsupervised slide representation learning. However, these approaches focus solely on the visual modality of patches, neglecting the rich semantic information embedded in textual data. In this work, we propose ProAlign, a cross-modal unsupervised slide representation learning framework. Specifically, we leverage a large language model (LLM) to generate descriptive text for the prototype types present in a WSI, introducing patch-text contrast to construct initial prototype embeddings. Furthermore, we propose a parameter-free attention aggregation strategy that utilizes the similarity between patches and these prototypes to form unsupervised slide embeddings applicable to a wide range of downstream tasks. Extensive experiments on four public datasets show that ProAlign outperforms existing unsupervised frameworks and achieves performance comparable to some weakly supervised models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge