Jiawen Li
To What Extent Do Token-Level Representations from Pathology Foundation Models Improve Dense Prediction?
Feb 03, 2026Abstract:Pathology foundation models (PFMs) have rapidly advanced and are becoming a common backbone for downstream clinical tasks, offering strong transferability across tissues and institutions. However, for dense prediction (e.g., segmentation), practical deployment still lacks a clear, reproducible understanding of how different PFMs behave across datasets and how adaptation choices affect performance and stability. We present PFM-DenseBench, a large-scale benchmark for dense pathology prediction, evaluating 17 PFMs across 18 public segmentation datasets. Under a unified protocol, we systematically assess PFMs with multiple adaptation and fine-tuning strategies, and derive insightful, practice-oriented findings on when and why different PFMs and tuning choices succeed or fail across heterogeneous datasets. We release containers, configs, and dataset cards to enable reproducible evaluation and informed PFM selection for real-world dense pathology tasks. Project Website: https://m4a1tastegood.github.io/PFM-DenseBench
HookMIL: Revisiting Context Modeling in Multiple Instance Learning for Computational Pathology
Dec 20, 2025Abstract:Multiple Instance Learning (MIL) has enabled weakly supervised analysis of whole-slide images (WSIs) in computational pathology. However, traditional MIL approaches often lose crucial contextual information, while transformer-based variants, though more expressive, suffer from quadratic complexity and redundant computations. To address these limitations, we propose HookMIL, a context-aware and computationally efficient MIL framework that leverages compact, learnable hook tokens for structured contextual aggregation. These tokens can be initialized from (i) key-patch visual features, (ii) text embeddings from vision-language pathology models, and (iii) spatially grounded features from spatial transcriptomics-vision models. This multimodal initialization enables Hook Tokens to incorporate rich textual and spatial priors, accelerating convergence and enhancing representation quality. During training, Hook tokens interact with instances through bidirectional attention with linear complexity. To further promote specialization, we introduce a Hook Diversity Loss that encourages each token to focus on distinct histopathological patterns. Additionally, a hook-to-hook communication mechanism refines contextual interactions while minimizing redundancy. Extensive experiments on four public pathology datasets demonstrate that HookMIL achieves state-of-the-art performance, with improved computational efficiency and interpretability. Codes are available at https://github.com/lingxitong/HookMIL.
StainNet: A Special Staining Self-Supervised Vision Transformer for Computational Pathology
Dec 11, 2025Abstract:Foundation models trained with self-supervised learning (SSL) on large-scale histological images have significantly accelerated the development of computational pathology. These models can serve as backbones for region-of-interest (ROI) image analysis or patch-level feature extractors in whole-slide images (WSIs) based on multiple instance learning (MIL). Existing pathology foundation models (PFMs) are typically pre-trained on Hematoxylin-Eosin (H&E) stained pathology images. However, images with special stains, such as immunohistochemistry, are also frequently used in clinical practice. PFMs pre-trained mainly on H\&E-stained images may be limited in clinical applications involving special stains. To address this issue, we propose StainNet, a specialized foundation model for special stains based on the vision transformer (ViT) architecture. StainNet adopts a self-distillation SSL approach and is trained on over 1.4 million patch images cropping from 20,231 publicly available special staining WSIs in the HISTAI database. To evaluate StainNet, we conduct experiments on an in-house slide-level liver malignancy classification task and two public ROI-level datasets to demonstrate its strong ability. We also perform few-ratio learning and retrieval evaluations, and compare StainNet with recently larger PFMs to further highlight its strengths. We have released the StainNet model weights at: https://huggingface.co/JWonderLand/StainNet.
ACT as Human: Multimodal Large Language Model Data Annotation with Critical Thinking
Nov 13, 2025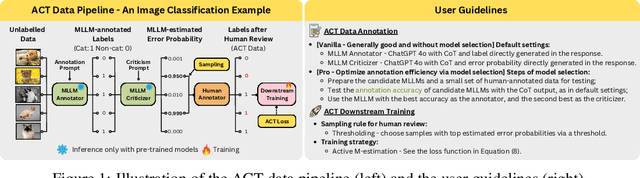



Abstract:Supervised learning relies on high-quality labeled data, but obtaining such data through human annotation is both expensive and time-consuming. Recent work explores using large language models (LLMs) for annotation, but LLM-generated labels still fall short of human-level quality. To address this problem, we propose the Annotation with Critical Thinking (ACT) data pipeline, where LLMs serve not only as annotators but also as judges to critically identify potential errors. Human effort is then directed towards reviewing only the most "suspicious" cases, significantly improving the human annotation efficiency. Our major contributions are as follows: (1) ACT is applicable to a wide range of domains, including natural language processing (NLP), computer vision (CV), and multimodal understanding, by leveraging multimodal-LLMs (MLLMs). (2) Through empirical studies, we derive 7 insights on how to enhance annotation quality while efficiently reducing the human cost, and then translate these findings into user-friendly guidelines. (3) We theoretically analyze how to modify the loss function so that models trained on ACT data achieve similar performance to those trained on fully human-annotated data. Our experiments show that the performance gap can be reduced to less than 2% on most benchmark datasets while saving up to 90% of human costs.
Deformable Attention Graph Representation Learning for Histopathology Whole Slide Image Analysis
Aug 07, 2025Abstract:Accurate classification of Whole Slide Images (WSIs) and Regions of Interest (ROIs) is a fundamental challenge in computational pathology. While mainstream approaches often adopt Multiple Instance Learning (MIL), they struggle to capture the spatial dependencies among tissue structures. Graph Neural Networks (GNNs) have emerged as a solution to model inter-instance relationships, yet most rely on static graph topologies and overlook the physical spatial positions of tissue patches. Moreover, conventional attention mechanisms lack specificity, limiting their ability to focus on structurally relevant regions. In this work, we propose a novel GNN framework with deformable attention for pathology image analysis. We construct a dynamic weighted directed graph based on patch features, where each node aggregates contextual information from its neighbors via attention-weighted edges. Specifically, we incorporate learnable spatial offsets informed by the real coordinates of each patch, enabling the model to adaptively attend to morphologically relevant regions across the slide. This design significantly enhances the contextual field while preserving spatial specificity. Our framework achieves state-of-the-art performance on four benchmark datasets (TCGA-COAD, BRACS, gastric intestinal metaplasia grading, and intestinal ROI classification), demonstrating the power of deformable attention in capturing complex spatial structures in WSIs and ROIs.
VGR: Visual Grounded Reasoning
Jun 16, 2025Abstract:In the field of multimodal chain-of-thought (CoT) reasoning, existing approaches predominantly rely on reasoning on pure language space, which inherently suffers from language bias and is largely confined to math or science domains. This narrow focus limits their ability to handle complex visual reasoning tasks that demand comprehensive understanding of image details. To address these limitations, this paper introduces VGR, a novel reasoning multimodal large language model (MLLM) with enhanced fine-grained visual perception capabilities. Unlike traditional MLLMs that answer the question or reasoning solely on the language space, our VGR first detects relevant regions that may help to solve problems, and then provides precise answers based on replayed image regions. To achieve this, we conduct a large-scale SFT dataset called VGR -SFT that contains reasoning data with mixed vision grounding and language deduction. The inference pipeline of VGR allows the model to choose bounding boxes for visual reference and a replay stage is introduced to integrates the corresponding regions into the reasoning process, enhancing multimodel comprehension. Experiments on the LLaVA-NeXT-7B baseline show that VGR achieves superior performance on multi-modal benchmarks requiring comprehensive image detail understanding. Compared to the baseline, VGR uses only 30\% of the image token count while delivering scores of +4.1 on MMStar, +7.1 on AI2D, and a +12.9 improvement on ChartQA.
Inverse Design of Metamaterials with Manufacturing-Guiding Spectrum-to-Structure Conditional Diffusion Model
Jun 08, 2025Abstract:Metamaterials are artificially engineered structures that manipulate electromagnetic waves, having optical properties absent in natural materials. Recently, machine learning for the inverse design of metamaterials has drawn attention. However, the highly nonlinear relationship between the metamaterial structures and optical behaviour, coupled with fabrication difficulties, poses challenges for using machine learning to design and manufacture complex metamaterials. Herein, we propose a general framework that implements customised spectrum-to-shape and size parameters to address one-to-many metamaterial inverse design problems using conditional diffusion models. Our method exhibits superior spectral prediction accuracy, generates a diverse range of patterns compared to other typical generative models, and offers valuable prior knowledge for manufacturing through the subsequent analysis of the diverse generated results, thereby facilitating the experimental fabrication of metamaterial designs. We demonstrate the efficacy of the proposed method by successfully designing and fabricating a free-form metamaterial with a tailored selective emission spectrum for thermal camouflage applications.
PathOrchestra: A Comprehensive Foundation Model for Computational Pathology with Over 100 Diverse Clinical-Grade Tasks
Mar 31, 2025Abstract:The complexity and variability inherent in high-resolution pathological images present significant challenges in computational pathology. While pathology foundation models leveraging AI have catalyzed transformative advancements, their development demands large-scale datasets, considerable storage capacity, and substantial computational resources. Furthermore, ensuring their clinical applicability and generalizability requires rigorous validation across a broad spectrum of clinical tasks. Here, we present PathOrchestra, a versatile pathology foundation model trained via self-supervised learning on a dataset comprising 300K pathological slides from 20 tissue and organ types across multiple centers. The model was rigorously evaluated on 112 clinical tasks using a combination of 61 private and 51 public datasets. These tasks encompass digital slide preprocessing, pan-cancer classification, lesion identification, multi-cancer subtype classification, biomarker assessment, gene expression prediction, and the generation of structured reports. PathOrchestra demonstrated exceptional performance across 27,755 WSIs and 9,415,729 ROIs, achieving over 0.950 accuracy in 47 tasks, including pan-cancer classification across various organs, lymphoma subtype diagnosis, and bladder cancer screening. Notably, it is the first model to generate structured reports for high-incidence colorectal cancer and diagnostically complex lymphoma-areas that are infrequently addressed by foundational models but hold immense clinical potential. Overall, PathOrchestra exemplifies the feasibility and efficacy of a large-scale, self-supervised pathology foundation model, validated across a broad range of clinical-grade tasks. Its high accuracy and reduced reliance on extensive data annotation underline its potential for clinical integration, offering a pathway toward more efficient and high-quality medical services.
Cross-Modal Prototype Allocation: Unsupervised Slide Representation Learning via Patch-Text Contrast in Computational Pathology
Mar 26, 2025Abstract:With the rapid advancement of pathology foundation models (FMs), the representation learning of whole slide images (WSIs) attracts increasing attention. Existing studies develop high-quality patch feature extractors and employ carefully designed aggregation schemes to derive slide-level representations. However, mainstream weakly supervised slide representation learning methods, primarily based on multiple instance learning (MIL), are tailored to specific downstream tasks, which limits their generalizability. To address this issue, some studies explore unsupervised slide representation learning. However, these approaches focus solely on the visual modality of patches, neglecting the rich semantic information embedded in textual data. In this work, we propose ProAlign, a cross-modal unsupervised slide representation learning framework. Specifically, we leverage a large language model (LLM) to generate descriptive text for the prototype types present in a WSI, introducing patch-text contrast to construct initial prototype embeddings. Furthermore, we propose a parameter-free attention aggregation strategy that utilizes the similarity between patches and these prototypes to form unsupervised slide embeddings applicable to a wide range of downstream tasks. Extensive experiments on four public datasets show that ProAlign outperforms existing unsupervised frameworks and achieves performance comparable to some weakly supervised models.
Multimodal Distillation-Driven Ensemble Learning for Long-Tailed Histopathology Whole Slide Images Analysis
Mar 02, 2025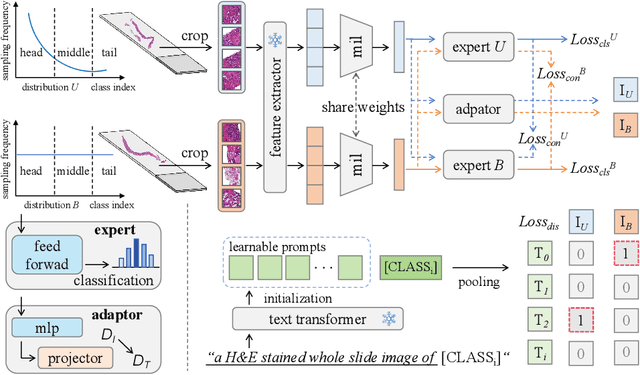


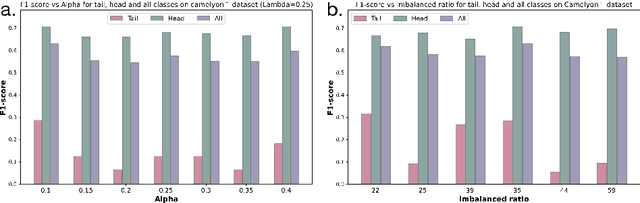
Abstract:Multiple Instance Learning (MIL) plays a significant role in computational pathology, enabling weakly supervised analysis of Whole Slide Image (WSI) datasets. The field of WSI analysis is confronted with a severe long-tailed distribution problem, which significantly impacts the performance of classifiers. Long-tailed distributions lead to class imbalance, where some classes have sparse samples while others are abundant, making it difficult for classifiers to accurately identify minority class samples. To address this issue, we propose an ensemble learning method based on MIL, which employs expert decoders with shared aggregators and consistency constraints to learn diverse distributions and reduce the impact of class imbalance on classifier performance. Moreover, we introduce a multimodal distillation framework that leverages text encoders pre-trained on pathology-text pairs to distill knowledge and guide the MIL aggregator in capturing stronger semantic features relevant to class information. To ensure flexibility, we use learnable prompts to guide the distillation process of the pre-trained text encoder, avoiding limitations imposed by specific prompts. Our method, MDE-MIL, integrates multiple expert branches focusing on specific data distributions to address long-tailed issues. Consistency control ensures generalization across classes. Multimodal distillation enhances feature extraction. Experiments on Camelyon+-LT and PANDA-LT datasets show it outperforms state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge